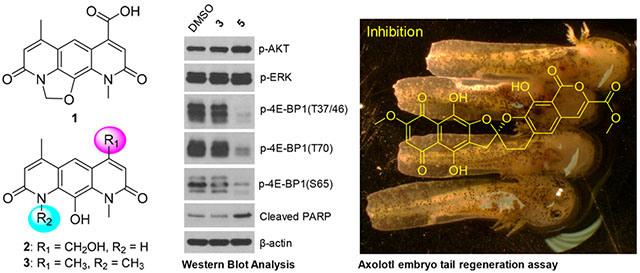
Abstract
We report the isolation and characterization of three new nybomycins (nybomycins B–D, 1–3) and six known compounds (nybomycin, 4; deoxynyboquinone, 5; α-rubromycin, 6; β-rubromycin, 7; γ-rubromycin, 8; and [2α(1E,3E),4β]-2-(1,3-pentadienyl)-4-piperidinol, 9) from the Rock Creek (McCreary County, KY) underground coal mine acid reclamation site isolate Streptomyces sp. AD-3-6. Nybomycin D (3) and deoxynyboquinone (5) displayed moderate (3) to potent (5) cancer cell line cytotoxicity and displayed weak to moderate anti-Gram-(+) bacterial activity, whereas rubromycins 6–8 displayed little to no cancer cell line cytotoxicity but moderate to potent anti-Gram-(+) bacterial and antifungal activity. Assessment of the impact of 3 or 5 cancer cell line treatment on 4E-BP1 phosphorylation, a predictive marker of ROS-mediated control of cap-dependent translation, also revealed deoxynyboquinone (5)-mediated downstream inhibition of 4E-BP1p. Evaluation of 1–9 in a recently established axolotl embryo tail regeneration assay also highlighted the prototypical telomerase inhibitor γ-rubromycin (8) as a new inhibitor of tail regeneration. Cumulatively, this work highlights an alternative nybomycin production strain, a small set of new nybomycin metabolites, and previously unknown functions of rubromycins (antifungal activity and inhibition of tail regeneration) and also provides a basis for revision of the previously proposed nybomycin biosynthetic pathway.
Graphical Abstract
INTRODUCTION
Nybomycins are pyridoquinolinedione-based metabolites of Streptomyces isolated from diverse environments including terrestrial soil samples,1,2 marine sediments,3,4 and the body surface of carpenter ants.5 These secondary metabolites are selective inhibitors of a quinoline-resistant Ser84Leu mutant DNA gyrase (GyrA), and synthetic deoxynybomycin analogues display good oral bioavailability, tolerance, and efficacy in murine infection models.6–8 Conversely, the development of nybomycin resistance surprisingly resensitizes a bacterium to quinolone antibiotics, and thus, nybomycins have been designated as “reverse antibiotics”.6,8 Nybomycins are also active against both proliferating and dormant Mycobacterium tuberculosis the proposed mechanistic basis for which is GyrA-independent.3 In addition, some nybomycins display potent cancer cell line cytotoxicity where NAD(P)H quinone oxidoreductase 1 (NQO1)-mediated bioreductive activation and corresponding intracellular reactive oxygen species (ROS) production are believed to be the driving factors.9–14 Nybomycin analogues can also inhibit cell division cycle 25 (CDC25) phosphatases that serve as key regulators of the eukaryotic cell cycle and are highly overexpressed in many cancers.15,16 Despite these prominent diverse biological functions of nybomycins, corresponding biosynthetic studies are limited to early metabolic labeling studies17,18 and the more recent annotation of a nybomycin biosynthetic gene cluster,4 both implicating 4-aminoanthranilic acid as a central progenitor. Given this context, access to new naturally occurring nybomycins, corresponding production strains, and/or their genomic-encoded biocatalysts may enable further probe/lead development and/or biosynthetic advances.
As part of an effort to discover new microbial natural products,19–28 their unique molecular targets,29,30 and/or corresponding biocatalysts,31 herein, we describe the isolation and structure elucidation of three new nybomycins (nybomycins B–D, 1–3) and six previously reported metabolites (nybomycin, 4; deoxynyboquinone, 5; α-rubromycin, 6; β-rubromycin, 7; γ-rubromycin, 8; and [2α(1E,3E),4β]-2-(1,3-pentadienyl)-4-piperidinol, 9) from the Rock Creek (McCreary County, KY) underground coal mine acid reclamation site isolate Streptomyces sp. AD-3-6. Bioactivity studies revealed compounds 3 and 5 to afford moderate (3) to potent (5) cancer cell line cytotoxicity and weak to moderate anti-Gram-(+) bacterial activity. In contrast, the isolated rubromycins displayed little to no cancer cell line cytotoxicity but moderate to potent anti-Gram-(+) bacterial and antifungal activity. Consistent with 5-mediated ROS production,9 and the recently established relationship between elevated [ROS] and the phosphorylation of the translational repressor 4E-BP1,29,30 5 also inhibited phosphorylation of 4E-BP1. Using an axolotl embryo tail regeneration assay,25,27,30,32,33 evaluation of the set of isolated metabolites also identified 8 as an inhibitor of tail regeneration. These cumulative studies highlight an alternative nybomycin production strain, a small set of new nybomycin metabolites, and previously unknown functions of rubromycins (antifungal activity and inhibition of tail regeneration). In addition, a revision of the previously proposed nybomycin biosynthetic pathway is put forth based on the new metabolites described.
RESULTS AND DISCUSSION
Six actinomycete strains were purified from a single soil sample collected near an acid mine reclamation site in McCreary County, Kentucky. Metabolic profiling of these strains implicated Streptomyces sp. AD-3-6 as capable of unique metabolite production based on an AntiBase 201734 database comparison. Scale-up fermentation (10 L) of Streptomyces sp. AD-3-6, followed by extraction, fractionation, and iterative chromatography (silica gel column chromatography, Sephadex LH-20 column chromatography and semipreparative C18 HPLC), gave nybomycins B (1, yield = 1.5 mg/L), C (2, yield = 0.5 mg/L), and D (3, yield = 0.3 mg/L), nybomycin (4, yield = 2.5 mg/L), deoxynyboquinone (5, yield = 0.2 mg/L), α-rubromycin (6, yield = 0.2 mg/L), β-rubromycin (7, yield = 1.1 mg/L), γ-rubromycin (8, yield = 0.6 mg/L), and [2α(1E,3E),4β]-2-(1,3-pentadienyl)-4-piperidinol (9, yield = 0.6 mg/L) (Supporting Information, Scheme S1).
Structure Elucidation.
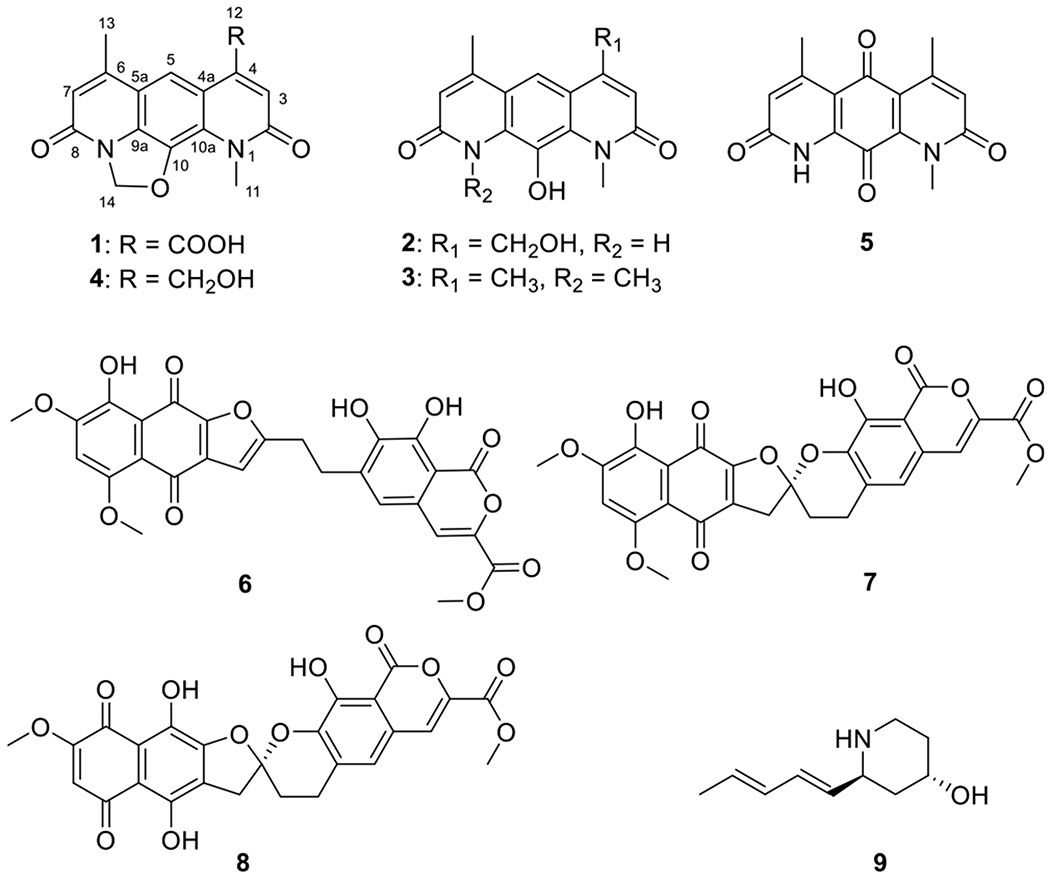
Compounds 1 (C16H12N2O5), 2 (C15H14N2O4), and 3 (C16H16N2O3) displayed UV–vis and NMR signatures consistent with previously reported nybomycins.1,2,7,9,35–37 1H NMR, 13C NMR, and HSQC data (Table 1) of 1 were consistent with the presence of 16 carbons, including two methyls [δH 4.01 (3H, s)/δC 33.8 (CH3-11), and δH 2.49 (3H, s)/δC 16.2 (CH3-13)], one methylene [δH 6.42 (2H, s)/δC 86.8 (CH2-14)], three sp2 methines [δH 7.47 (1H, s)/δC 122.5 (CH-3), δH 8.59 (1H, s)/δC 116.7 (CH-5), and δH 6.78 (1H, s)/δC 117.8 (CH-7)], seven sp2 nonprotonated carbons [δC 140.2 (C-4), δC 118.8 (C-4a), δC 117.1 (C-5a), δC 155.0 (C-6), δC 130.9 (C-9a), δC 138.2 (C-10), and δC 124.7 (C-10a)], and three acid/amide carbonyls [δC 163.3 (C-2), δC 160.0 (C-8), and δC 169.0 (C-12)]. Key HMBC correlations [from CH3-11 to C-2/C-10a, from CH3-13 to C-5a/C-6, from H-3 to C-2/C-4/C-4a/C-11/C-12, from H-5 to C-4/C-4a/C-6/C-9a/C-10a, and from H-7 to C-5a/C-8/C-13] further highlighted the close structural relationship of 1 and nybomycin (4, a previously reported metabolite also produced by Streptomyces sp. AD-3-6)1,35 with structural divergence in the C-4 substitution (Chart 1). Specifically, in 1, the prototypical nybomycin C-4 exocyclic CH2OH has been replaced by a COOH, key support for which was the observed CH-3 (δH 7.47) to C-12 (δC 169.0) HMBC correlation. Thus, the structure of 1 was established as a new member of the nybomycin family and subsequently named nybomycin B.
Table 1.
13C (100 MHz) and 1H (400 MHz) NMR Spectroscopic Data of Nybomycins B–D (1–3) and Nybomycin 4 in CF3COOD
| nybomycin B (1) |
nybomycin C (2) |
nybomycin D (3) |
nybomycin (4) |
|||||
|---|---|---|---|---|---|---|---|---|
| no. | δC, type | δH, mult. | δC, type | δH, mult. | δC, type | δH, mult. | δC, type | δH, mult. |
| 2 | 163.3, C | 163.8, C | 164.8, C | 162.8, C | ||||
| 3 | 122.5, CH | 7.47, s | 116.2, CH | 7.42, s | 115.1, CH | 7.30, s | 110.3, CH | 7.30, s |
| 4 | 140.2, C | 146.5, C | 156.3, C | 155.0, C | ||||
| 4a | 118.8, C | 119.0, C | 123.2, C | 119.0, C | ||||
| 5 | 116.7, CH | 8.59, s | 115.9, CH | 8.35, s | 116.5, CH | 8.41, s | 111.9, CH | 8.41, s |
| 5a | 117.1, C | 120.1, C | 123.2, C | 116.4, C | ||||
| 6 | 155.0, C | 156.8, C | 156.3, C | 151.3, C | ||||
| 7 | 117.8, CH | 6.78, s | 116.4, CH | 115.1, CH | 119.2, CH | |||
| 8 | 160.0, C | 162.7, C | 164.8, C | 159.4, C | ||||
| 9a | 130.9, C | 132.1, C | 136.9, C | 131.1, C | ||||
| 10 | 138.2, C | 132.2, C | 133.6, C | 137.8, C | ||||
| 10a | 124.7, C | 131.8, C | 136.9, C | 122.4, C | ||||
| 11 | 33.8, CH3 | 4.01, s | 36.0, CH3 | 4.43, s | 38.4, CH3 | 4.37, s | 33.4, CH3 | 4.09, s |
| 12 | 169.0, C | 64.3, CH2 | 5.99, s | 17.8, CH3 | 2.86, s | 59.4, CH3 | 5.09, s | |
| 13 | 16.2, CH3 | 2.49, s | 17.5, CH3 | 2.91, s | 17.8, CH3 | 2.86, s | 15.5, CH3 | 2.50, s |
| 14 | 86.8, CH2 | 6.42, s | 38.4, CH3 | 4.37, s | 85.9, CH2 | 6.42, s | ||
Chart 1.
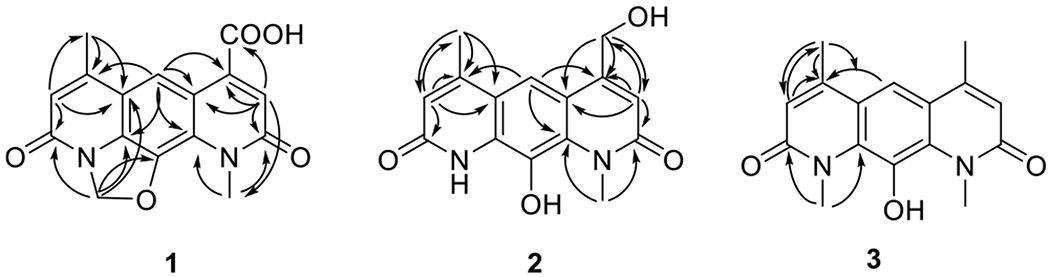
Cumulative analysis of NMR data (Table 1) indicated both 2 and 3 also shared the pyrido[3,2-g]quinoline-2,8(1H,9H)-dione core common to 1 and 4. Compound 2 was distinguished by the lack of the 4 dihydro-oxazole CH2. In contrast, the C-4 and N-9 methyl substitutions were the sole differentiating features of 3 (supported by HMBC correlations from CH3-9/CH3-1 to C-8/C-2 and C-9a/C-10a, from CH-13/CH3-12 to C-6/C-4, C-7/C-4, and C-5a/C-4a), indicating the similarity of ring A/C in compound 3 (Chart 1, Figure 1, Table 1). Metabolites 2 and 3 were thereby designated as new nybomycins C and D.
Figure 1.
Selected HMBC (→) correlations of nybomycins B–D (1–3).
Comparison of the NMR and MS data with literature values established the remaining known compounds as nybomycin (4),35 deoxynyboquinone (5),9 α-rubromycin (6),38 β-rubromycin (7),39 γ-rubromycin (8),40 and [2α(1E,3E),4β]-2-(1,3-pentadienyl)-4-piperidinol (9).41
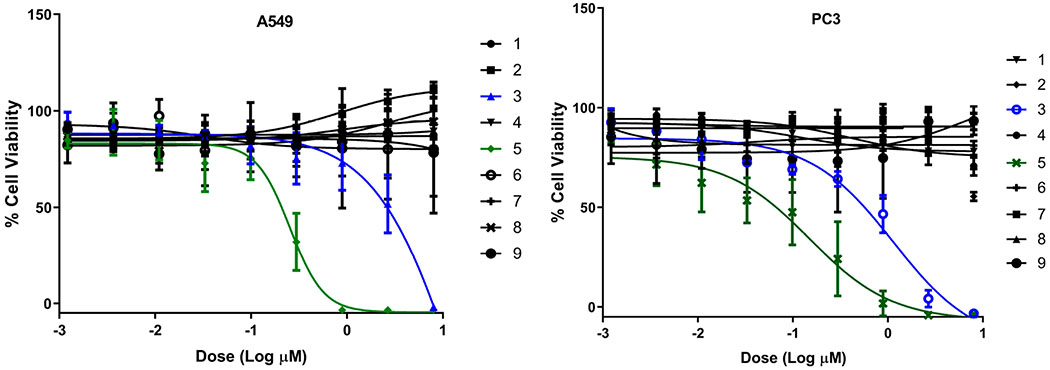
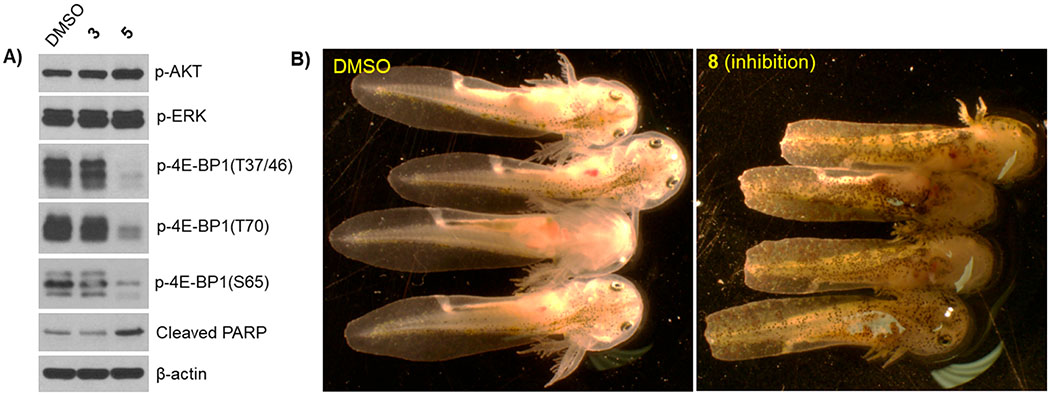
All isolated compounds (1–9) were evaluated in standard antibacterial, antifungal, and cancer cell line cytotoxicity assays (Table 2, Figure 2, and Supporting Information, Figure S2). Nybomycin D (3) and deoxynyboquinone (5) were the only two metabolites to display moderate (3) to potent (5) cancer cell line cytotoxicity [A549 (human non-small cell lung): EC50 for 3 (15.17 μM) and 5 (0.25, μM); PC3 (human prostate): EC50 for compounds 3 (1.14 μM) and 5 (0.15 μM)]. Subsequent evaluation of 4E-BP1 phosphorylation, a predictive marker of ROS-mediated effects,29,30 in HCT116 (human colorectal) cancer cells treated with 1 μM 3 or 5 revealed selective inhibition of 4E-BP1p in the presence of 5 (Figure 3A). Nybomycins 3 and 5 also displayed weak to moderate anti-Gram-(+) bacterial activity. In contrast, rubromycins 6–8 displayed little to no cancer cell line cytotoxicity but moderate to potent anti-Gram-(+) bacterial and antifungal activity. Evaluation of all isolated metabolites 1–9 in our recently developed axolotl embryo tail regeneration assay25,27,30,32,33 also highlighted 8 as a new inhibitor of tail regeneration (Figure 3B and Supporting Information, Figure S3).
Table 2.
Antimicrobial and Cytotoxic Activities of Compounds 1–9a
| MIC μM (μg/mL) | EC50 μM | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| compounds | S. aureus | M. luteus | B. subtilis | M. aurum | E. coli | S. enterica | S. cerevisae | A549 | PC3 |
| 1 | >24 (7.5) | >24 (7.5) | >24 (7.5) | >24 (7.5) | >24 (7.5) | >24 (7.5) | >24 (7.5) | >100 | >100 |
| 2 | >24 (6.9) | >24 (6.9) | >24 (6.9) | >24 (6.9) | >24 (6.9) | >24 (6.9) | >24 (6.9) | >100 | >100 |
| 3 | 12 (3.4) | 12 (3.4) | 6 (1.7) | >24 (6.8) | >24 (6.8) | >24 (6.8) | >24 (6.8) | 15.17 | 1.14 |
| 4 | >24 (7.2) | 12 (3.6) | 12 (3.6) | >24 (7.2) | >24 (7.2) | >24 (7.2) | >24 (7.2) | >100 | >100 |
| 5 | 12 (3.4) | 12 (3.4) | 12 (3.4) | >24 (6.8) | >24 (6.8) | >24 (6.8) | >24 (6.8) | 0.25 | 0.15 |
| 6 | 3 (1.6) | 1.5 (0.8) | 0.4 (0.2) | >24 (12.9) | >24 (12.9) | >24 (12.9) | >24 (12.9) | >100 | >100 |
| 7 | 12 (6.4) | 6 (3.2) | 12 (6.4) | >24 (12.9) | >24 (12.9) | >24 (12.9) | 12 (6.4) | >100 | >100 |
| 8 | 0.075 (0.04) | 0.15 (0.08) | 0.02 (0.01) | 12 (6.3) | >24 (12.6) | >24 (12.6) | 3 (1.6) | >100 | >100 |
| 9 | >24 (4.0) | >24 (4.0) | >24 (4.0) | >24 (4.0) | >24 (4.0) | >24 (4.0) | >24 (4.0) | >100 | >100 |
Antibacterial/antifungal MIC values were obtained after 16–48 h incubation. Kanamycin [S. aureus, 7.5 (4); M. luteus, 15 (9); B. subtilis, 2 (1); S. enterica, 14 (8); and E. coli 7.5 (4)], Ampicillin [S. aureus, 12 (4); M. luteus, 25 (9); S. enterica, 25 (9); and E. coli 25 (9)] rifampicin [M. aurum, 2 (1)], amphotericin B [S. cerevisiae, 2 (1)] were used as positive controls. Cytotoxicity EC50/IC50 values were obtained after 72 h incubation. Actinomycin D and H2O2 [A549 (non-small cell lung), PC3 (prostate) human cancer cell lines] were used as positive control at 20 μM and 2 mM concentration, respectively (0% viable cells).
Figure 2.
Dose–response of compounds 1–9 against A549 (non-small cell lung) and PC3 (prostate) human cancer cell lines (72 h). For EC50 values, see Table 2.
Figure 3.
(A) HCT116 cells were treated with 2 μM 3 and 1 μM 5 or DMSO (negative control) for 6 h followed by Western blot analysis for the indicated proteins. (B) Impact of 1 μM 8 on axolotl embryo tail regeneration at 7 dpa compared to vehicle control (DMSO). Note that control embryos but not embryos treated with 8 regenerated a rounded tail tip typical of the unamputated condition.
Discussion.
Including the new naturally occurring nybomycins B–D (1–3) reported herein, 21 naturally occurring metabolites reminiscent of pyridoquinolinedione-based nybomycins have been reported.34 These include nybomycin (4, produced by Streptomyces sp.),1,2,35 hydroxynybomycin (derived from Streptomyces sp. D57),42 deoxynybomycin (derived from S. hyalinum and marine Streptomyces sp. B8855),37 deoxynyboquinone (derived from the deep-sea actinomycete Pseudonocardia sp. SCSIO 01299),9,43 and other closely related metabolites BE-12233 [produced by Streptomyces sp. BA-12233 (FERM P-10492)],44 Sch-538415 (produced by Streptomyces sp.),45,46 pseudonocardians A–C (derived from the deep-sea actinomycete Pseudonocardia sp. SCSIO 01299),43 diazaquinomycin A [also known as antibiotic OM 704A and NSC 626554; derived from Streptomyces sp. om-704-ka 333 (FERM-p 6520) and Streptomyces sp. GW48/1497],47–49 diazaquinomycin B (also known as antibiotic OM 704B and 9,10-dihydrodiazaquinomycin A; produced by Streptomyces sp. om-704),47,49 diazaquinomycins C–D (produced by Streptomyces sp. GW48/1497),48 and diazaquinomycins E–G (produced by the marine-derived Streptomyces sp.).50–52 The current study extends this previous work with both additional insights regarding metabolite function and biosynthesis.
From a functional perspective, the current work offers the following two primary advances. First, although NAD(P)H quinone oxidoreductase 1 (NQO1)-mediated bioreductive activation of 5 and corresponding intracellular ROS production is well-precedented,9 this study is the first to demonstrate the corresponding effect of 5 on 4E-BP1 phosphorylation. The phosphorylation status of 4E-BP1 was recently implicated as a potential predictive marker in response to ROS-based anticancer agents29,30. Dysregulation of cap-dependent translation through redundant phosphorylation of the translational repressor 4E-BP1 by multiple oncogenic pathways, such as PI3K/AKT/mTOR and RAS/RAF/MEK/ ERK, is associated with malignant progression and therapeutic resistance53–60. Compounds that induce H2O2 and/or ROS production, such as 5, activate the peroxisome-bound tuberous sclerosis complex, which leads to subsequent inhibition of mTORC1-mediated 4E-BP1 phosphorylation and concomitant repression of cap-dependent translation and cancer cell/tumor growth29,61,62. Second, this study is also the first to reveal γ-rubromycin (8) as an inhibitor of axolotl embryo tail regeneration. In addition to their ability to inhibit HIV reverse transcriptase,63–66, both β- and γ-rubromycin (7 and 8, respectively) are also prototypical inhibitors of telomerare63,66–69. Telomerase function is a central player in regeneration, aging, and cancer, where inhibition of telomerase correlates with reduced pluripotency/plasticity, premature aging, and reduced lifespan in various models70–74. Importantly, axolotl telomerase increases during axolotl neonate tail regeneration based on Western blotting.75 Although additional mechanistic studies are lacking, the ability of 8 to inhibit axolotl embryo tail regeneration implicates 8 as new chemical biology tool to mechanistically interrogate this unique regenerative model further.
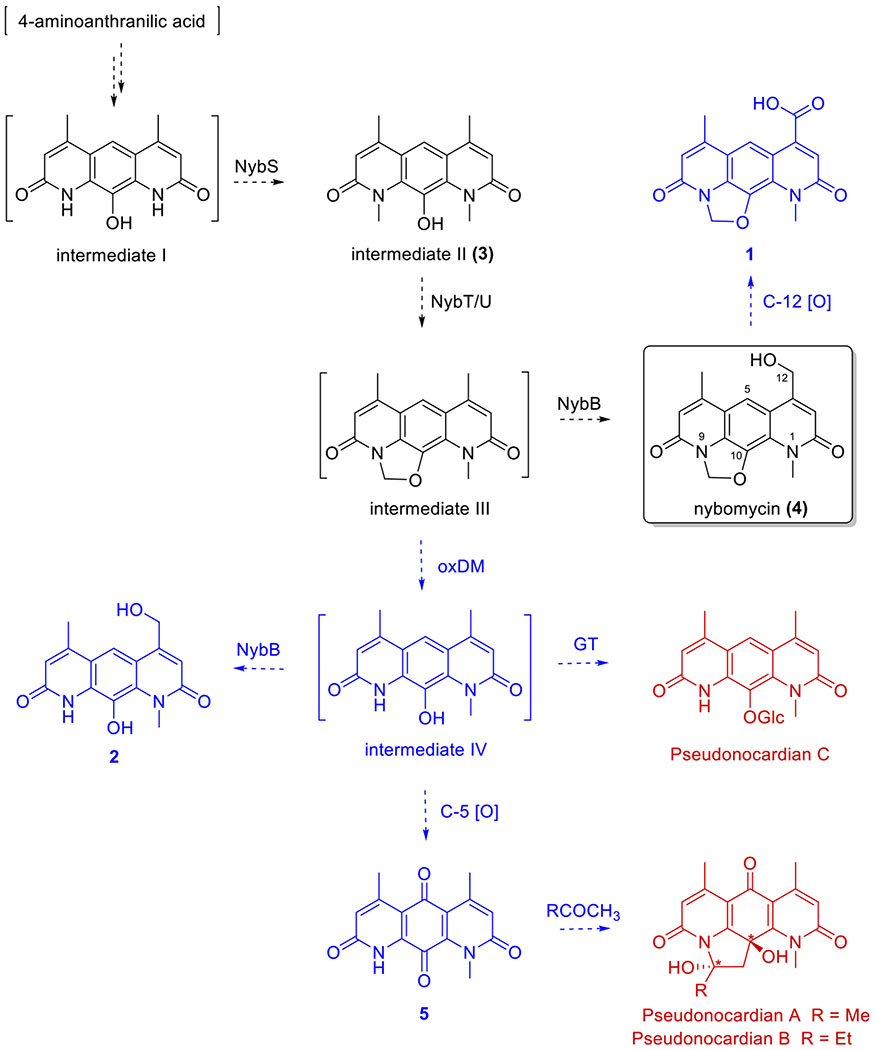
From a biosynthetic standpoint, the current work provides additional support for the biosynthetic pathway recently put forth by Luzhetskyy et al. and implicates new potential enzymatic and/or chemical avenues for diversification of the 4 scaffold (Figure 4).4 The central pyridoquinolinedione core of nybomycin was previously proposed to derive from 4-aminoanthracilic acid followed by NybS-catalyzed N-1/9-dimethylation to generate 3. The isolation of 3 from Streptomyces sp. AD-3-6 described herein provides the first direct support of 3 as a probable biosynthetic intermediate. Subsequent NybT/U-catalyzed isopenicillin N synthase-like ring closure was proposed to afford hypothetical intermediate III, followed by NybB-catalyzed C-12 hydroxylation to 4. Within this context, additional C-12 oxidation of 4 would yield 1, one of the major pyridoquinolinedione-based metabolites isolated in the current study. The isolation of metabolites 2 and 5, both lacking the methylene bridge, suggests a central C-14 oxidative demethylation step potentially originating from putative intermediate III. As a chemical precedent, previously reported chemical degradation of a semisynthetic intermediate III using MnO2 yielded intermediate IV.76 Following demethylation, intermediate IV could be hydroxylated by NybB to give 2, oxidized to the p-quinone 5, or glycosylated to form pseudonocardian C, the latter two of which are previously reported metabolites of Pseudonocardia sp. SCSIO 01299.43 Metabolite 5 could also serve as the precursor for pseudonocardins A and B, wherein the pyrrolidine ring may surprisingly reflect an artifact due to metabolite isolation via acetone or 2-butanone extraction. Consistent with this hypotheses, incubation of 5 in the presence of acetone (Supporting Information, Figure S36), followed by LC-MS analysis, gave a product with a UV/vis and MS signature consistent with that of pseudonocardin A. Cumulatively, the new metabolites and concepts resulting from this study are anticipated to enable future biochemical studies that support or refute the proposed biosynthetic hypotheses highlighted above.
Figure 4.
Streptomyces sp. AD-3-6-derived nybomycins 2–5 within the context of the biosynthetic pathway put forth by Luzhetskyy and colleagues (black) and pseudonocardians previously reported by Zhang et al. (red; *only relative stereochemistry assigned). Proposed biosynthetic revisions based on the current study are highlighted (blue). Compounds in brackets are putative intermediates, and dashed arrows represent putative transformations with possible enzyme catalysts noted where possible. Metabolite 3 reported herein was previously proposed by Luzhetskyy et al. as a biosynthetic intermediate, whereas metabolite 5 and pseudonocardians were previously reported by Zhang et al.
MATERIALS AND METHODS
General Experimental Procedures.
UV spectra were recorded on an Ultrospec 8000 spectrometer (GE, Pittsburgh, PA, USA). All NMR spectra were recorded at 400 MHz for 1H and 100 MHz for 13C with Varian Inova NMR spectrometers (Agilent, Santa Clara, CA, USA). HPLC-MS was conducted with an Agilent 6120 quadrupole MSD mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) equipped with an Agilent 1200 series quaternary LC system and an Eclipse XDB-C18 column (150 × 4.6 mm, 5 μm). HR-ESI-MS spectra were recorded on an AB SCIEX Triple TOF 5600 system (AB Sciex, Framingham, MA, USA). HPLC analyses were performed on an Agilent 1260 system equipped with a photodiode array (PDA) detector and a Phenomenex C18 column (250 × 4.6 mm, 5 μm; Phenomenex, Torrance, CA). Semipreparative HPLC separation was performed on a Varian Prostar 210 HPLC system equipped with a PDA detector using a Supelco DiscoveryBio wide pore C18 column (250 × 21.2 mm, 10 μm; flow rate, 8 mL/min; Sigma-Aldrich, St. Louis, MO, USA). Size exclusion chromatography was performed on Sephadex LH-20 (25–100 μm; GE Healthcare, Piscataway, NJ, USA). Amberlite XAD16N resin (20–60 mesh) was purchased from Sigma-Aldrich (St. Louis, MO, USA). TLC silica gel plates (60 F254) were purchased from EMD Chemicals Inc. (Darmstadt, Germany). Staphylococcus aureus, Bacillus subtilis, Salmonella enterica, Mycobacterium aurum, and Saccharomyces cerevisiae strains and human cancer cell lines A549 (ATCC CCL185 human lung non-small cell carcinoma), HCT116 (ATCC CCL-247 human colorectal carcinoma), and PC3 (ATCC CRL1435 human prostate adenocarcinoma) were obtained from ATCC (Manassas, VA, USA). Micrococcus luteus and Escherichia coli strains were obtained from NRRL (Peoria, IL, USA). All solvents used were of ACS grade and purchased from the Pharmco-AAPER (Brookfield, CT, USA). All other reagents used were reagent grade and purchased from Sigma-Aldrich (St. Louis, MO, USA).
Isolation and Identification of Streptomyces sp. AD-3-6.
The soil sample was collected from an acid mine drainage site in McCreary County, KY (GPS coordinates: 36°42′03.4″N 84°34′04.7″W). Metabolic profiling and strain isolation followed previously reported protocols.19,20,22 Strain identification, based on 16S rRNA sequencing following previously described protocols,26,27 revealed 99% identity (BLAST search) with the 16S rRNA gene sequence of Streptomyces iakyrus strain NBRC 13401. The sequence of 16S rRNA has been deposited in the NCBI nucleotide database with the accession number KX902490.
Scale-up Fermentation.
Streptomyces sp. AD-3-6 was cultivated in three 250 mL Erlenmeyer flasks, each containing 50 mL of medium A (soluble starch, 20.0 g/L; glucose, 10.0 g/L; peptone, 5.0 g/L; yeast extract, 5.0 g/L; NaCl, 4.0 g/L; K2HPO4, 0.5 g/L; MgSO4·7H2O, 0.5 g/L; CaCO3, 2.0 g/L, pH 7.0). After 3 days of incubation at 28 °C with 200 rpm agitation, the cultures were used to inoculate 100 flasks (250 mL), each containing 100 mL of medium A (total 10 L). The fermentation was continued for 10 days at 28 °C with 200 rpm agitation. The combined culture broth was centrifuged at 3000g for 30 min (4 °C) to provide the solid biomass and supernatant. The cumulative biomass (mycelium) was extracted with MeOH (3 × 600 mL), and the corresponding recovered organics were subsequently evaporated in vacuo at 40 °C to yield 15.3 g of crude extract. The supernatant was mixed with 3% (w/v) XAD-16 resin and stirred overnight, followed by filtration. The resin was washed with H2O (3 × 600 mL) and then extracted with MeOH until the eluant was colorless. The combined MeOH extracts were subsequently evaporated to afford 10.8 g of crude extract. Both extracts (obtained from the biomass and supernatant) revealed a similar metabolite profile based on HPLC and TLC analyses and were therefore combined.
As highlighted in Scheme S1, the combined crude extract (26.1 g) was subjected to HP-20SS resin column chromatography (800g, 40 × 8 cm) eluted with a gradient of aqueous MeOH (20–100%) to yield five fractions (A–E). Fraction B (1.5 g) was resolved by size-exclusion chromatography (Sephadex LH-20, 4 × 100 cm, 2 mL/min, MeOH) to yield three subfractions, B1–B3 (100 mL each). Subfraction B2 (0.3 g) was further purified by a semipreparative HPLC (10–30% CH3CN/0.025% TFA over 30 min) to afford compound 1 (15.0 mg, white amorphous powder, retention time: 11.8 min). Fraction C (1.3 g) was subjected to Sephadex LH-20 column (4 × 100 cm, 2 mL/min, MeOH) and the recovered subfraction C2 (0.2 g) further purified by a semipreparative HPLC (5–40% CH3CN/0.025% TFA over 30 min) to yield compounds 2 (5.0 mg, white amorphous powder, retention time: 10.1 min) and 9 [2-(1,3-pentadien-1-yl)-4-piperidinol, 6.0 mg, white amorphous powder, retention time: 13.4 min]. Fraction D (2.7 g) was subjected to Sephadex LH-20 column chromtography (4 × 100 cm, 2 mL/min, MeOH) to yield three subfractions, D1–D3 (120 mL each). Subfraction D2 (0.8 g) was further purified by a semipreparative HPLC (20–45% CH3CN/0.025% TFA over 25 min) to yield compounds 3 (3.0 mg, white amorphous powder, retention time: 14.5 min), 5 (2.0 mg, white amorphous powder, retention time: 13.4 min), and 4 (25.0 mg, white amorphous powder, retention time: 12.0 min). Fraction E (2.4 g) was subjected to a silica gel column (50 g, 12 × 4 cm) eluted with CHCl3/MeOH (10:0–1:1) to yield five fractions, E1–E5. Subfraction E5 (0.3 g) was further purified by semipreparative HPLC (45–75% CH3CN/0.05% TFA over 30 min) to afford compound 6 (2.0 mg, red amorphous powder, retention time: 21.6 min). Subfraction E3 (0.4 g) was further purified by semipreparative HPLC (50–70% CH3CN/0.05% TFA over 25 min) to afford compounds 7 (11.0 mg, red amorphous powder, retention time: 21.0 min) and 8 (6.0 mg, red amorphous powder, retention time: 24.1 min).
Nybomycin B (1): white amorphous powder; UV (DMSO) λmax (log ε) 273 (9.27), 294 (8.04), 379 (3.66) nm; for 13C and 1H NMR data, see Table 1; (+)-ESI-MS m/z 313.0 [M + H]+; (+)-HR-ESI-MS m/z 313.0820 [M + H]+ (calcd for C16H13N2O5, 313.0824).
Nybomycin C (2): white amorphous powder; UV (DMSO) λmax (log ε) 284 (15.37), 357 (4.77), 375 (5.57) nm; for 13C and 1H NMR data, see Table 1; (+)-ESI-MS m/z 287.0 [M + H]+, (−)-ESI-MS m/z 285.0 [M − H]−; (+)-HR-ESI-MS m/z 287.1030 [M + H]+ (calcd for C15H15N2O4, 287.1032).
Nybomycin D (3): white amorphous powder; UV (DMSO) λmax (log ε) 290 (14.44), 359 (3.61), 376 (3.67) nm; for 13C and 1H NMR data, see Table 1; (+)-ESI-MS m/z 285.1 [M + H]+, (−)-ESI-MS m/z 283.0 [M − H]−; (+)-HR-ESI-MS m/z 285.1240 [M + H]+ (calcd for C16H17N2O3, 285.1239).
Antibacterial, Antifungal, and Cancer Cell Line Viability Assays.
Antibacterial (Staphylococcus aureus ATCC 6538, Micrococcus luteus ATCC 15307, Bacillus subtilis ATCC 6633, Mycobacterium aurum ATCC 23366, Escherichia coli ATCC 12435, and Salmonella enterica ATCC 10708), antifungal (Saccharomyces cerevisiae ATCC 204508), and cytotoxicity (human non-small cell lung cancer cell A549 and prostate cancer cell PC3) assays were accomplished in triplicate following our previously reported protocols.20,77 Antibacterial/antifungal MIC values were obtained after 16–48 h incubation. Vehicle (DMSO) was used as the negative control, and kanamycin and ampicillin (S. aureus, M. luteus, B. subtilis, M. aurum, S. enterica, and E. coli), amphotericin B (S. cerevisiae), and actinomycin D (A549 and PC3) were used as positive controls.
Axolotl Embryo Tail Regeneration Assay.
The axolotl embryo tail regeneration assay was conducted following our previously reported protocols.25,27,30,32,33 Axolotls (RRID:AGSC_100E) were obtained from the Ambystoma Genetic Stock Center (RRID:SCR_006372). Vehicle (DMSO) was used as the negative control, and the Hsp90 inhibitor geldanamycin was used as a positive control.
Western Blot Analysis.
Western blot analysis was performed as previously described.29,30
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grant Nos. R24 OD21479 (S.R.V., J.S.T.), R01 CA203257 (Q.B.S., J.S.T.), R01 CA175105 (Q.B.S.), and R01 GM115261 (J.S.T.), the University of Kentucky College of Pharmacy, the University of Kentucky Markey Cancer Center, and the National Center for Advancing Translational Sciences (UL1TR000117 and UL1TR001998). We thank the College of Pharmacy NMR Center (University of Kentucky) for NMR support.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jnatprod.9b01015.
The workup isolation scheme, axolotl embryo tail regeneration assay figures treated with compounds 1–9, and full spectroscopic data (NMR and mass) of compounds 1–9 (PDF)
The authors declare the following competing financial interest(s): J.S.T. is a co-founder of Centrose (Madison, WI).
REFERENCES
- (1).Strelitz F; Flon H; Asheshov IN Proc. Natl. Acad. Sci. U. S. A 1955, 41, 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Eble TE; Boyack GA; Large CM; Devries WH Antibiot. Chemother 1958, 8, 627–630. [PubMed] [Google Scholar]
- (3).Arai M; Kamiya K; Pruksakorn P; Sumii Y; Kotoku N; Joubert JP; Moodley P; Han C; Shin D; Kobayashi M Bioorg. Med. Chem 2015, 23, 3534–3541. [DOI] [PubMed] [Google Scholar]
- (4).Rodríguez Estévez M; Myronovskyi M; Gummerlich N; Nadmid S; Luzhetskyy A Mar. Drugs 2018, 16, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zakalyukina YV; Birykov MV; Lukianov DA; Shiriaev DI ; Komarova ES; Skvortsov DA; Kostyukevich Y; Tashlitsky VN; Polshakov VI; Nikolaev E; Sergiev PV; Osterman IA Biochimie 2019, 160, 93–99. [DOI] [PubMed] [Google Scholar]
- (6).Hiramatsu K; Igarashi M; Morimoto Y; Baba T; Umekita M; Akamatsu Y Int. J. Antimicrob. Agents 2012, 39, 478–485. [DOI] [PubMed] [Google Scholar]
- (7).Parkinson EI; Bair JS; Nakamura BA; Lee HY; Kuttab HI; Southgate EH; Lezmi S; Lau GW; Hergenrother P Nat. Commun 2015, 6, 6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Morimoto Y; Baba T; Sasaki T; Hiramatsu K Int. J. Antimicrob. Agents 2015, 46, 666–673. [DOI] [PubMed] [Google Scholar]
- (9).Bair JS; Palchaudhuri R; Hergenrother PJ J. Am. Chem. Soc 2010, 132, 5469–5478. [DOI] [PubMed] [Google Scholar]
- (10).Parkinson EI; Hergenrother PJ Acc. Chem. Res 2015, 48, 2715–2723. [DOI] [PubMed] [Google Scholar]
- (11).Ponnuraj N; Kolossov V; Parkinson E; Benefiel A; Hergenrother P; Gaskins H FASEB J. 2014, 28, 655.10. [Google Scholar]
- (12).Lee HY; Parkinson EI; Granchi C; Paterni I; Panigrahy D; Seth P; Minutolo F; Hergenrother PJ ACS Chem. Biol 2017, 12, 1416–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Parkinson EI; Bair JS; Cismesia M; Hergenrother PJ ACS Chem. Biol 2013, 8, 2173–2183. [DOI] [PubMed] [Google Scholar]
- (14).Huang X; Dong Y; Bey EA; Kilgore JA; Bair JS; Li LS; Patel M; Parkinson EI; Wang Y; Williams NS; Gao J; Hergenrother PJ; Boothman DA Cancer Res. 2012, 72, 3038–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Nussbaum F; Ebbinghaus A; Mayer-Bartschmid A; Zitzmann W; Wiese W-B; Stadler M; Anlauf S CDC25 inhibitors. Patent Appl. EP2130831A1, 2009.
- (16).Sur S; Agrawal DK Mol. Cell. Biochem 2016, 416, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Nadzan AM; Rinehart KL Jr. J. Am. Chem. Soc 1976, 98, 5012–5014. [DOI] [PubMed] [Google Scholar]
- (18).Nadzan AM; Rinehart KL Jr. J. Am. Chem. Soc 1977, 99, 4647–4654. [DOI] [PubMed] [Google Scholar]
- (19).Shaaban KA; Wang X; Elshahawi SI; Ponomareva LV; Sunkara M; Copley GC; Hower JC; Morris AJ; Kharel MK; Thorson JS J. Nat. Prod 2013, 76, 1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wang X; Shaaban KA; Elshahawi SI; Ponomareva LV; Sunkara M; Zhang Y; Copley GC; Hower JC; Morris AJ; Kharel MK; Thorson JS J. Nat. Prod 2013, 76, 1441–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wang X; Elshahawi SI; Shaaban KA; Fang L; Ponomareva LV; Zhang Y; Copley GC; Hower JC; Zhan CG; Kharel MK; Thorson JS Org. Lett 2014, 16, 456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang X; Shaaban KA; Elshahawi SI; Ponomareva LV; Sunkara M; Copley GC; Hower JC; Morris AJ; Kharel MK; Thorson JS J. Antibiot 2014, 67, 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wang X; Reynolds AR; Elshahawi SI; Shaaban KA; Ponomareva LV; Saunders MA; Elgumati IS; Zhang Y; Copley GC; Hower JC; Sunkara M; Morris AJ; Kharel MK; Van Lanen SG; Prendergast MA; Thorson JS Org. Lett 2015, 17, 2796–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Shaaban KA; Saunders MA; Zhang Y; Tran T; Elshahawi SI; Ponomareva LV; Wang X; Zhang J; Copley GC; Sunkara M; Kharel MK; Morris AJ; Hower JC; Tremblay MS; Prendergast MA; Thorson JS J. Nat. Prod 2017, 80, 2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wang X; Zhang Y; Ponomareva LV; Qiu Q; Woodcock R; Elshahawi SI; Chen X; Zhou Z; Hatcher BE; Hower JC; Zhan CG; Parkin S; Kharel MK; Voss SR; Shaaban KA; Thorson JS Angew. Chem. Int. Ed 2017, 56, 2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Abbas M; Elshahawi SI; Wang X; Ponomareva LV; Sajid I; Shaaban KA; Thorson JS J. Nat. Prod 2018, 81, 2560–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wang X; Abbas M; Zhang Y; Elshahawi SI; Ponomareva LV; Cui Z; Van Lanen SG; Sajid I; Voss SR; Shaaban KA; Thorson JS J. Nat. Prod 2019, 82, 1686–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Fatima A; Aftab U; Shaaban KA; Thorson JS; Sajid I BMC Microbiol. 2019, 19 (1), 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ye Q; Zhang Y; Cao Y; Wang X; Guo Y; Chen J; Horn J ; Ponomareva LV; Chaiswing L; Shaaban KA; Wei Q; Anderson BD; St Clair DK; Zhu H; Leggas M; Thorson JS; She QB Cell. Chem. Biol 2019, 26, 366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zhang Y; Ye Q; Ponomareva LV; Cao Y; Liu Y; Cui Z; Van Lanen SG; Voss SR; She QB; Thorson JS Chem. Sci 2019, 10, 7641–7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Elshahawi SI; Cao H; Shaaban KA; Ponomareva LV; Subramanian T; Farman ML; Spielmann HP; Phillips GN Jr.; Thorson JS; Singh S Nat. Chem. Biol 2017, 13, 366–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ponomareva LV; Athippozhy A; Thorson JS; Voss SR Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol 2015, 178, 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Voss SR; Ponomareva LV; Dwaraka VB; Pardue KE; Baddar N; Rodgers AK; Woodcock MR; Qiu Q; Crowner A; Blichmann D; Khatri S; Thorson JS Sci. Rep 2019, 9, 6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Laatsch H AntiBase 2017; Wiley-VCH: Weinheim, Germany, 2017. [Google Scholar]
- (35).Rinehart KL Jr.; Leadbetter G; Larson RA; Forbis RM J. Am. Chem. Soc 1970, 92, 6994–6995. [DOI] [PubMed] [Google Scholar]
- (36).Forbis RM; Rinehart KL Jr. J. Am. Chem. Soc 1970, 92, 6995–6996. [DOI] [PubMed] [Google Scholar]
- (37).Naganawa H; Wakashiro T; Yagi A; Kondo S; Takita T; Hamada M; Maeda K; Umezawa H J. Antibiot. 1970, 23, 365–368. [DOI] [PubMed] [Google Scholar]
- (38).Brockmann H; Lenk W; Schwantje G; Zeeck A Tetrahedron Lett. 1966, 7, 3525–3530. [DOI] [PubMed] [Google Scholar]
- (39).Puder C; Loya S; Hizi A; Zeeck A Eur. J. Org. Chem 2000, 2000, 729–735. [Google Scholar]
- (40).Akai S; Kakiguchi K; Nakamura Y; Kuriwaki I; Dohi T; Harada S; Kubo O; Morita N; Kita Y Angew. Chem. Int. Ed 2007, 46, 7458–7461. [DOI] [PubMed] [Google Scholar]
- (41).Grabley S; Hammann P; Kluge H; Wink J; Kricke P; Zeeck A J. Antibiot. 1991, 44, 797–800. [DOI] [PubMed] [Google Scholar]
- (42).Nadzan AM; Rinehart KL Jr.; Sokolski WT J. Antibiot 1977, 30, 523–524. [DOI] [PubMed] [Google Scholar]
- (43).Li S; Tian X; Niu S; Zhang W; Chen Y; Zhang H; Yang X; Zhang W; Li W; Zhang S; Ju J; Zhang C Mar. Drugs 2011, 9, 1428–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ojiri K; Suda H; Okura A; Kawamura K; Okanishi M Anticancer agent BE-12233 manufacture with Streptomyces. Patent Appl. JP 03030688 A2, 1991.
- (45).Chu M; Mierzwa R; Xu L; Yang SW; He L; Patel M; Stafford J; Macinga D; Black T; Chan TM; Gullo V Bioorg. Med. Chem. Lett 2003, 13, 3827–3829. [DOI] [PubMed] [Google Scholar]
- (46).Pettit GR; Du J; Pettit RK; Richert LA; Hogan F; Mukku VJ; Hoard MS J. Nat. Prod 2006, 69, 804–806. [DOI] [PubMed] [Google Scholar]
- (47).Murata M; Miyasaka T; Tanaka H; Omura S J. Antibiot 1985, 38, 1025–1033. [DOI] [PubMed] [Google Scholar]
- (48).Maskey RP; Grün-Wollny I; Laatsch H Nat. Prod. Res 2005, 19, 137–142. [DOI] [PubMed] [Google Scholar]
- (49).Omura S; Iwai Y; Hinotozawa K; Tanaka H; Takahashi Y; Nakagawa A J. Antibiot. 1982, 35, 1425–1429. [DOI] [PubMed] [Google Scholar]
- (50).Mullowney M; O’hAinmhire E; Shaikh A; Wei X; Tanouye U; Santarsiero B; Burdette J; Murphy B Mar. Drugs 2014, 12, 3574–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Prior AM; Sun D RSC Adv. 2019, 9, 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Mullowney MW; Hwang CH; Newsome AG; Wei X; Tanouye U; Wan B; Carlson S; Barranis NJ; Ó hAinmhire E; Chen W-L; Krishnamoorthy K; White J; Blair R; Lee H; Burdette JE; Rathod PK; Parish T; Cho S; Franzblau SG; Murphy BT ACS Infect. Dis 2015, 1, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Bhat M; Robichaud N; Hulea L; Sonenberg N; Pelletier J; Topisirovic I Nat. Rev. Drug Discovery 2015, 14, 261–278. [DOI] [PubMed] [Google Scholar]
- (54).Boussemart L; Malka-Mahieu H; Girault I; Allard D; Hemmingsson O; Tomasic G; Thomas M; Basmadjian C; Ribeiro N; Thuaud F; Mateus C; Routier E; Kamsu-Kom N; Agoussi S; Eggermont AM; Desaubry L; Robert C; Vagner S Nature 2014, 513, 105–109. [DOI] [PubMed] [Google Scholar]
- (55).Cai W; Ye Q; She QB Oncotarget 2014, 5, 6015–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Ducker GS; Atreya CE; Simko JP; Hom YK; Matli MR; Benes CH; Hann B; Nakakura EK; Bergsland EK; Donner DB; Settleman J; Shokat KM; Warren RS. Oncogene 2014, 33, 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Hsieh AC; Costa M; Zollo O; Davis C; Feldman ME; Testa JR; Meyuhas O; Shokat KM; Ruggero D Cancer Cell 2010, 17, 249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Mi W; Ye Q; Liu S; She QB Oncotarget 2015, 6, 13962–13977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).She QB; Halilovic E; Ye Q; Zhen W; Shirasawa S; Sasazuki T; Solit DB; Rosen N Cancer Cell 2010, 18, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ye Q; Cai W; Zheng Y; Evers BM; She QB Oncogene 2014, 33, 1828–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Zhang J; Kim J; Alexander A; Cai S; Tripathi DN; Dere R; Tee AR; Tait-Mulder J; Di Nardo A; Han JM; Kwiatkowski E ; Dunlop EA; Dodd KM; Folkerth RD; Faust PL; Kastan MB; Sahin M; Walker CL Nat. Cell Biol 2013, 15, 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Chen MF; Keng PC; Shau H; Wu CT; Hu YC; Liao SK; Chen WC Int. J. Radiat. Oncol., Biol., Phys 2006, 64, 581–591. [DOI] [PubMed] [Google Scholar]
- (63).Ueno T; Takahashi H; Oda M; Mizunuma M; Yokoyama A; Goto Y; Mizushina Y; Sakaguchi K; Hayashi H Biochemistry 2000, 39, 5995–6002. [DOI] [PubMed] [Google Scholar]
- (64).Goldman ME; Salituro GS; Bowen JA; Williamson JM; Zink DL; Schleif WA; Emini EA Mol. Pharmacol 1990, 38, 20–25. [PubMed] [Google Scholar]
- (65).Bernardo CE; Silva PJ PeerJ 2014, 2, e470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Atkinson DJ; Brimble MA Nat. Prod. Rep 2015, 32, 811–840. [DOI] [PubMed] [Google Scholar]
- (67).Cohn EP; Wu KL; Pettus TR; Reich NO J. Med. Chem 2012, 55, 3678–3686. [DOI] [PubMed] [Google Scholar]
- (68).Mizushina Y; Takeuchi T; Sugawara F; Yoshida H Mini-Rev. Med. Chem 2012, 12, 1135–1143. [DOI] [PubMed] [Google Scholar]
- (69).Chen JL-Y; Sperry J; Ip NY; Brimble MA MedChemComm 2011, 2, 229–245. [Google Scholar]
- (70).West MD; Sternberg H; Labat I; Janus J; Chapman KB; Malik NN; de Grey AD; Larocca D Regener. Med 2019, 14, 867–886. [DOI] [PubMed] [Google Scholar]
- (71).Godic A Clin. Dermatol 2019, 37, 320–325. [DOI] [PubMed] [Google Scholar]
- (72).Kumar M; Lechel A; Gunes C Genes 2016, 7, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Carneiro MC; de Castro IP; Ferreira MG Dis. Models & Mech 2016, 9, 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Wu RA; Upton HE; Vogan JM; Collins K Annu. Rev. Biochem 2017, 86, 439–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Alibardi L J. Exp. Zool, Part A 2015, 323, 757–766. [DOI] [PubMed] [Google Scholar]
- (76).Knoll WM; Huxtable RJ; Rinehart KL Jr. J. Am. Chem. Soc 1973, 95, 2703–2704. [DOI] [PubMed] [Google Scholar]
- (77).Shaaban KA; Elshahawi SI; Wang X; Horn J; Kharel MK; Leggas M; Thorson JS J. Nat. Prod 2015, 78, 1723–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.