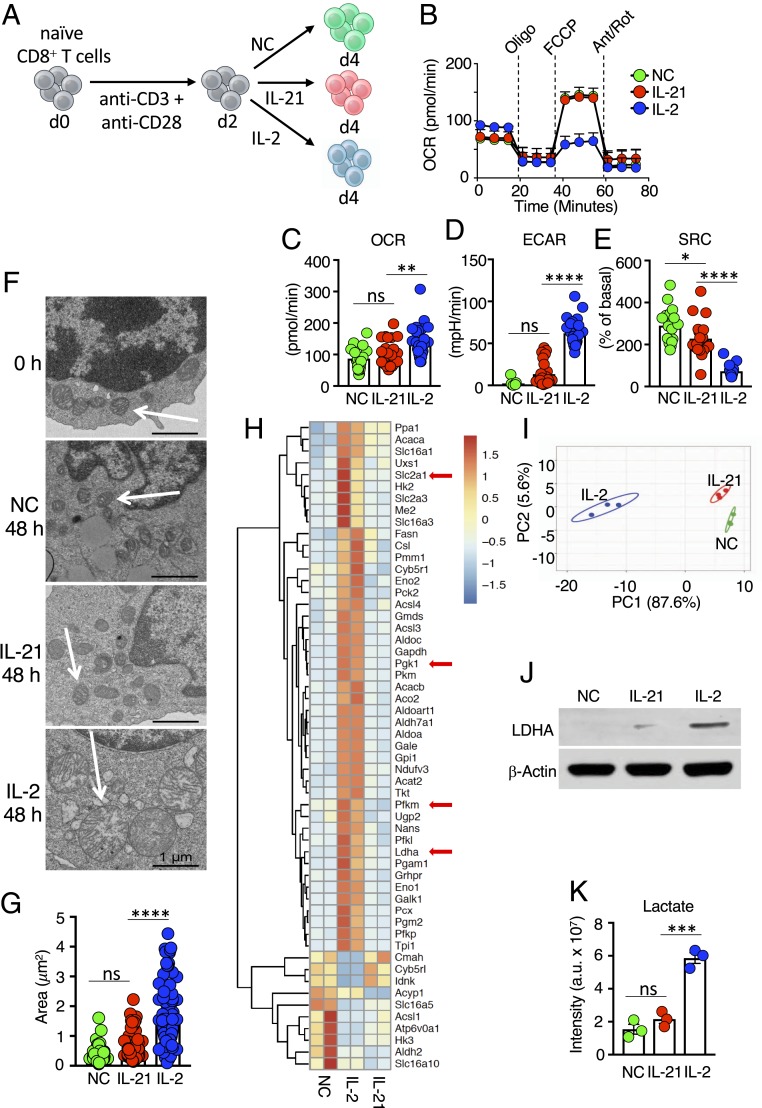
Fig. 1.
Distinctive metabolic effects of IL-2 versus IL-21. (A) Schematic of protocol for stimulating CD8+ T cells. Naive T cells were isolated, activated with anti-CD3 and anti-CD28 for 2 d, and cultured with NC, IL-21, or IL-2 for another 2 d. (B) Seahorse experiment measuring the OCR for CD8+ T cells treated with NC, IL-21, or IL-2 following treatment with oligomycin, FCCP, and antimycin A/rotenone. Data are representative of 11 independent experiments. (C–E) Bar graphs indicating basal OCR (C), ECAR (D), and SRC (E), as measured from Seahorse assays. (F) Electron micrographs are representative images from two independent experiments. (Scale bar: 1 μm.) (G) Calculated area of mitochondria from electron micrographs from cells treated with IL-2, IL-21, or NC. (H) RNA-Seq heatmap of differentially expressed metabolic genes from cells treated with NC, IL-2, or IL-21. Shown is the scale for fold induction or repression. (I) Principal component analysis plot based on metabolomics data of cells treated with NC, IL-2, or IL-21. (J) Western blotting of LDHA protein. (K) LC-MS–based analysis of intracellular lactate. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.