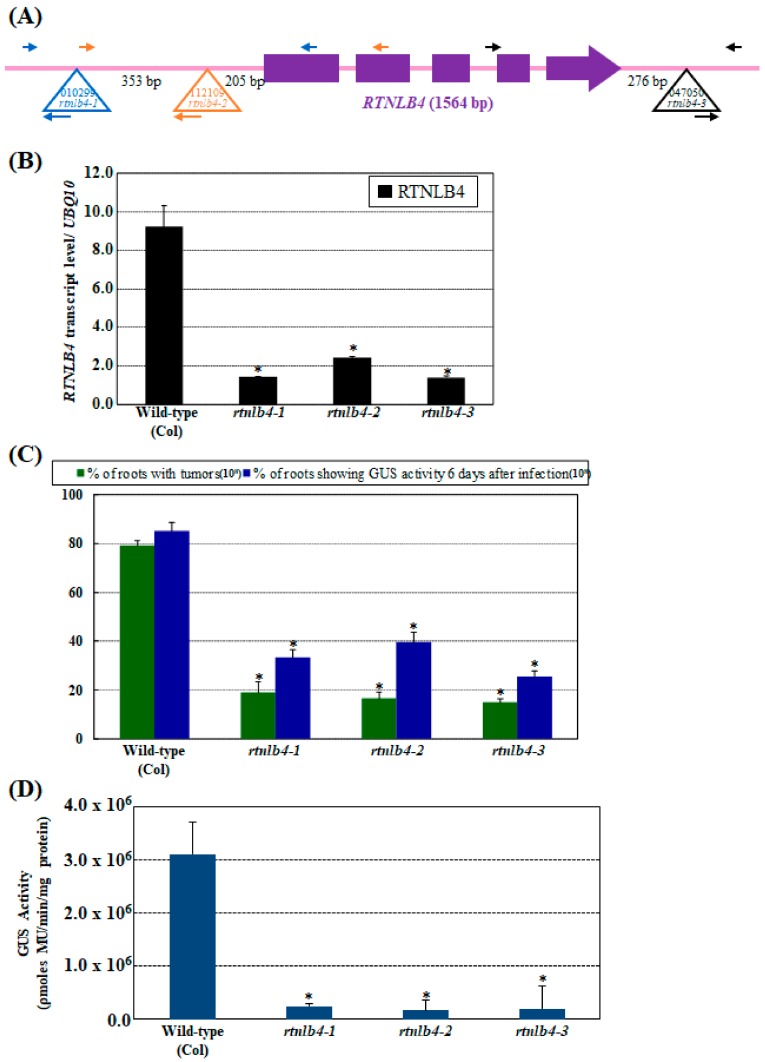
Figure 1.
The Arabidopsis rtnlb4 T-DNA insertion mutants were recalcitrant to Agrobacterium tumefaciens transformation. (A) Schematic representations of the T-DNA insertion regions around the Arabidopsis RTNLB4 gene. Purple boxes represent exon regions of the RTNLB4 gene. The large open triangle represents T-DNA insertion sites in the RTNLB4 gene. The long and short arrows indicate the locations of primers used in genomic DNA PCR analysis. (B) qPCR results of the RTNLB4 transcript in rtnlb4-1, rtnlb4-2, and rtnlb4-3 mutants. UBQ10 (polyubiquitin 10) transcript level was an internal control. Data are mean ± SE from at least three PCR reactions of each mutant. (C) Transformation efficiencies of three rtnlb4 mutant lines and wild-type plants. Green bars indicate percentages of root segments forming tumors at 1 month after infection with 108 cfu mL−1 tumorigenic A. tumefaciens A208 strain. Blue bars show percentages of root segments with GUS activity 6 days after infection with 108 cfu mL−1 A. tumefaciens At849 strain. Data are mean ± SE from more than 15 plants. At least 80 root segments were examined for each plant. (D) Seedlings from three rtnlb4 mutant lines showed reduced susceptibility to transient transformation. Mutant seedlings were infected with 107 cfu mL−1 acetosyringone (AS)-treated A. tumefaciens strain for 3 days to determine transient transformation efficiencies. Data are mean ± SE. * p < 0.05 compared with the wild-type by pairwise Student’s t test.