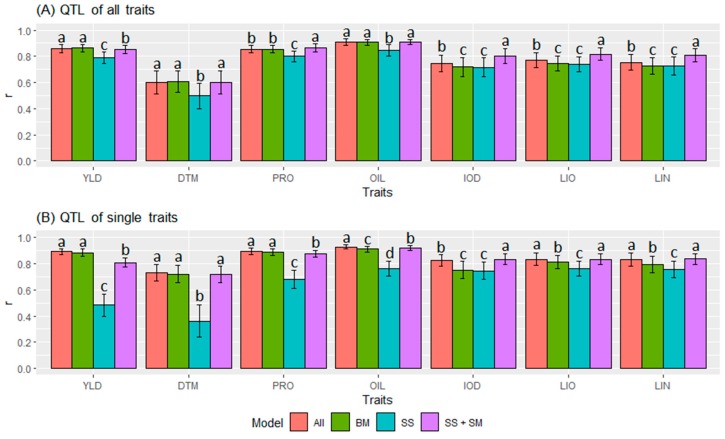
Figure 8.
Comparisons of genomic prediction accuracy (r ± s) by different statistical models, including SNP-based single-locus model (SS), SS+SNP-based multi-locus model (SM), haplotype-block-based model (BM), and all three models of SS+SM+BM (All), which were used for quantitative trait locus (QTL) identification. (A) QTL of all traits were used for GS, and (B) QTL of single traits were used for GS. A tag quantitative trait nucleotide (QTN) for each QTL was used for analyses. For each trait, different letters represent statistical significance of r values among different GWAS models. YLD, seed yield; DTM, days to maturity; PRO, protein content; OIL, oil content; IOD, iodine value; LIO, linoleic acid content (LIO); LIN, linolenic acid content; SNP, single nucleotide polymorphism.