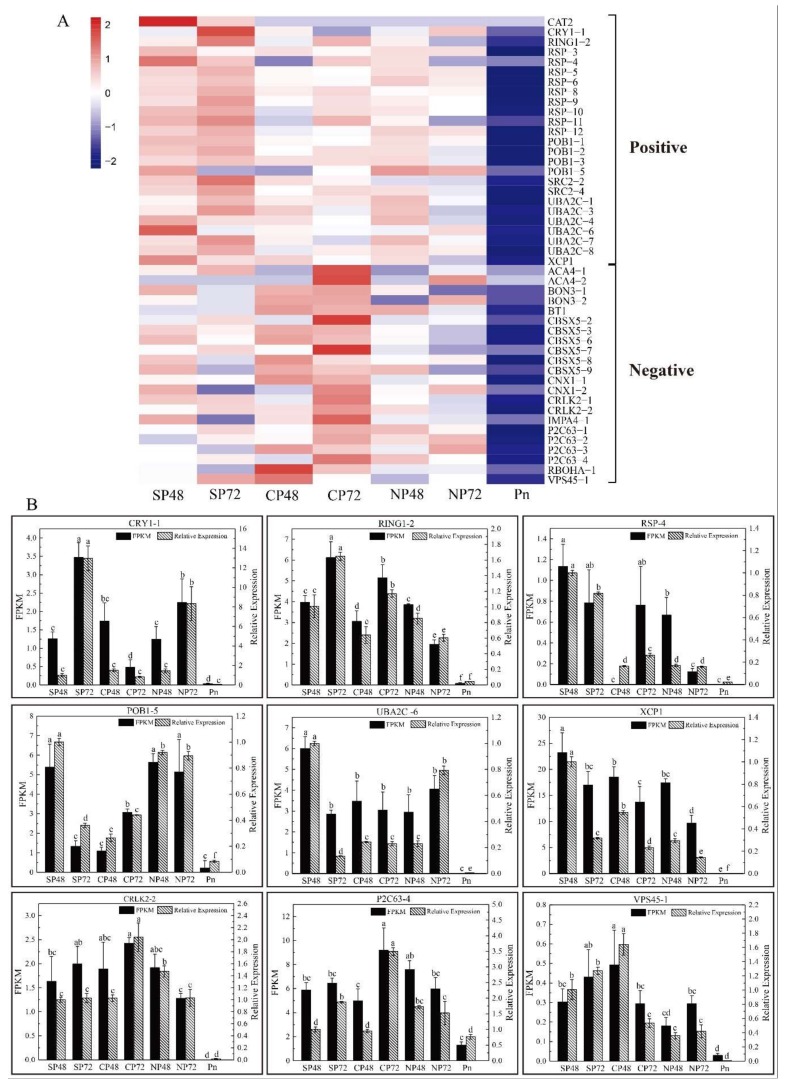
Figure 5.
Expression patterns of programmed cell death (PCD)-related genes in self-, cross- and non-pollinated pistils and mature pollen of Camellia oleifera. (A) The heatmap of PCD-related genes in self-, cross- and non-pollinated pistils and mature pollen. Positive refers to positive-regulated transcripts. Negative refers to negative-regulated transcripts. SP48/72, self-pollinated pistils at 48 h/72 h; CP48/72, cross-pollinated pistils at 48 h/72 h; NP48/72, non-pollinated pistils at 48 h/72 h and Pn, mature pollen. CAT2, catalase isozyme 2; CRY1, cryptochrome-1; RING1, E3 ubiquitin-protein ligase RING1; RSP, CO(2)-response secreted protease; SRC2, SRC2 homolog; UBA2C, UBP1-associated protein 2C; XCP1, xylem cysteine proteinase1; ACA4, alpha carbonic anhydrase 4; BON3, BONZAI 3; BT1, adenine nucleotide transporter BT1; CBSX5, CBS domain-containing protein CBSX5; CNX1, molybdopterin adenylyltransferase; CRLK2, leucine-rich repeat receptor-like serine/threonine-protein kinase CRLK2; IMPA4, importin subunit alpha-4; P2C63, phosphatase 2C 63; RBOHA, respiratory burst oxidase homolog protein A and VPS45, vacuolar protein sorting-associated protein 45. (B) Expression level of CRY1-1, RING1-2, RSP-4, POB1, UBA2C-6, XCP1, CRLK2-2, P2C63-4 and VPS45-1 based on FPKM and qRT-PCR data. FPKM = fragments per kilobase per million mapped fragments. SP48 sample was the reference to calculate the relative expression data. Error bars indicate SD. Different letters represent significant difference at mRNA levels (p < 0.05).