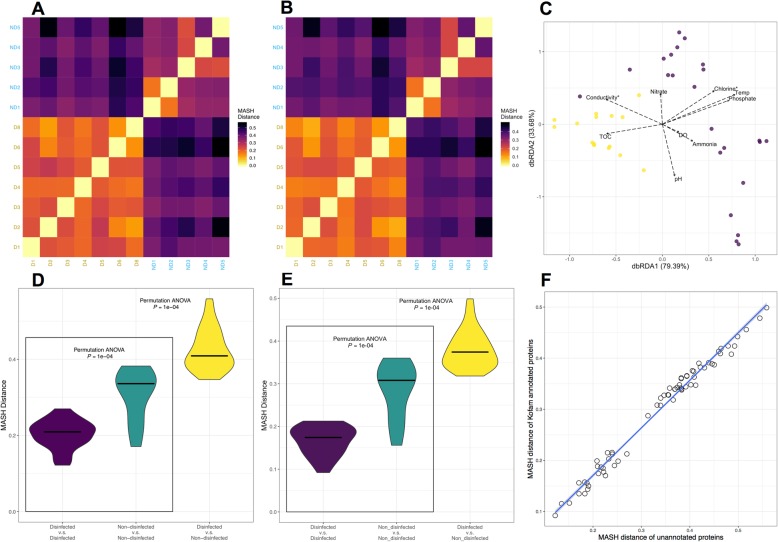
Fig. 4.
Comparison of functional potential among all and KEGG protein-coding amino acid sequences across sampling locations. This analysis estimates dissimilarity in amino acid composition of samples, similar to the nucleotide composition analyses presented earlier. a, b Heatmaps based on pairwise Mash distances of all protein-coding sequences and Bray-Curtis distances using KO. Heatmaps are colored according to Mash/Bray-Curtis distance; yellow denotes a distance of 0. Labels on x- and y-axis are colored according to disinfection strategies; dark golden denotes samples with chlorine, while blue denotes samples without disinfectant residuals. c MDS clustering of using Bray-Curtis distances using KO abundances between samples with vectors representing water chemistry/environmental parameters implemented using dbRDA. Violin plots indicating the distribution of pairwise d Mash distances of all and e Bray-Curtis distances KEGG-annotated proteins. Crossbars indicate the median value of Mash distances. f Correlation between pairwise Mash distances estimated using all and Bray-Curtis distances for KEGG-annotated protein coding sequences