Abstract
Fusarium oxysporum is the most economically important and commonly encountered species of Fusarium. This soil-borne fungus is known to harbour both pathogenic (plant, animal and human) and non-pathogenic strains. However, in its current concept F. oxysporum is a species complex consisting of numerous cryptic species. Identification and naming these cryptic species is complicated by multiple subspecific classification systems and the lack of living ex-type material to serve as basic reference point for phylogenetic inference. Therefore, to advance and stabilise the taxonomic position of F. oxysporum as a species and allow naming of the multiple cryptic species recognised in this species complex, an epitype is designated for F. oxysporum. Using multi-locus phylogenetic inference and subtle morphological differences with the newly established epitype of F. oxysporum as reference point, 15 cryptic taxa are resolved in this study and described as species.
Keywords: cryptic species, diversity, human and plant pathogens, species complex, subspecific classification
INTRODUCTION
Fusarium oxysporum is the most economically important and commonly encountered species of Fusarium. This soil-borne asexual fungus is known to harbour both pathogenic (plant, animal and human) and non-pathogenic strains (Leslie & Summerell 2006) and is also ranked fifth on a list of top 10 fungal pathogens based on scientific and economic importance (Dean et al. 2012, Geiser et al. 2013). Historically, F. oxysporum has been defined by the asexual phenotype as no sexual morph has yet been discovered, even though several studies have indicated the possible presence of a cryptic sexual cycle (Arie et al. 2000, Yun et al. 2000, Aoki et al. 2014, Gordon 2017). This is further supported by phylogenetic studies that place F. oxysporum within the Gibberella Clade (Baayen et al. 2000, O’Donnell et al. 2009, 2013). These studies also showed that F. oxysporum displays a complicated phylogenetic substructure, indicative of multiple cryptic species within F. oxysporum (Gordon & Martyn 1997, Laurence et al. 2014). As with other Fusarium species complexes, the F. oxysporum species complex (FOSC) has suffered from multiple taxonomic/classification systems applied in the past.
Diederich F.L. von Schlechtendal first introduced F. oxysporum in 1824, isolated from a rotten potato tuber (Solanum tuberosum) collected in Berlin, Germany. Wollenweber (1913) placed F. oxysporum within the section Elegans along with eight other Fusarium species and numerous varieties and forms based on similarity of the micro- and macroconidial morphology and dimensions. Snyder & Hansen (1940) later consolidated and reduced all species within the section Elegans into F. oxysporum and designated 25 special forms (formae speciales) within this species. These special forms were further expanded on by Gordon (1965) to 66, most of which are still used in literature today.
The use of special forms or formae speciales as subspecific rank in F. oxysporum classification has become common practice due to the broad morphological delineation of this species (Leslie & Summerell 2006). This informal subspecific rank is defined based on the plant pathogenicity of the particular F. oxysporum strain and excludes both clinical and non-pathogenic strains (Armstrong & Armstrong 1981, Gordon & Martyn 1997, Kistler 1997, Baayen et al. 2000, Leslie & Summerell 2006). Therefore, F. oxysporum strains attacking the same plant host are generally considered to belong to the same special form. Although this homologous trait has led to erroneous assumptions considering a specific special form to be phylogenetically monophyletic, several studies (O’Donnell et al. 1998, 2004, 2009, O’Donnell & Cigelnik 1999, Baayen et al. 2000, Lievens et al. 2009b, Van Dam et al. 2016) have highlighted the para- and polyphyletic relationships within several F. oxysporum special forms, e.g., F. oxysporum f. sp. batatas, F. oxysporum f. sp. cubense and F. oxysporum f. sp. vasinfectum. Additionally, several F. oxysporum special forms are able to infect and cause disease in more than one (sometimes unrelated) plant hosts, whereas others are highly specialised to a specific plant host (Armstrong & Armstrong 1981, Gordon & Martyn 1997, Kistler 1997, Baayen et al. 2000, Leslie & Summerell 2006, Fourie et al. 2011).
Naming F. oxysporum special forms are not subject to the International Code of Nomenclature for algae, fungi, and plants (ICN; McNeill et al. 2012, Thurland et al. 2018), and therefore no diagnosis (in Latin and/or English), nor the deposit of type material in a recognised repository is required. This decision was made due to the difficulty in accepting special forms within the Code, even though these strains are of great importance to plant pathologists and breeders (Deighton et al. 1962, Gordon 1965, Armstrong & Armstrong 1981). Several studies on F. oxysporum indicate that between 70 to over 150 special forms are known in F. oxysporum (Booth 1971, Armstrong & Armstrong 1981, Kistler 1997, Baayen et al. 2000, Leslie & Summerell 2006, Lievens et al. 2008, O’Donnell et al. 2009, Fourie et al. 2011, Laurence et al. 2014, Gordon 2017). At present Index Fungorum (http://www.indexfungorum.org/) lists 124 special forms in F. oxysporum, whereas MycoBank (http://www.mycobank.org/) list 127 special forms. Further careful scrutiny of literature revealed that 144 special forms have been named until February 2018 (Table 1). Although the special forms concept of Snyder & Hansen (1940) is still applied today, additional subspecific classification systems for special forms of F. oxysporum have also been introduced, which include haplotypes, races and vegetative compatibility groups (VCGs).
Table 1.
List of known special forms of Fusarium oxysporum.
The haplotype subspecific classification system was introduced by Chang et al. (2006) and later expanded upon by O’Donnell et al. (2008, 2009) to include strains from both the FOSC and Neocosmospora (formerly the F. solani (FSSC) species complex). This classification system is based on unique multilocus genotypes within the species complex, aimed to resolve communication problems among public health and agricultural scientists (O’Donnell et al. 2008). Chang et al. (2006) proposed a standardised haplotype nomenclature system that depict the species complex, species and genotype. O’Donnell et al. (2009) was able to identify 256 unique two-locus haplotypes from 850 isolates representing 68 special forms of F. oxysporum as well as environmental and clinical strains. However, this classification system is not in common use as a reference, and a continuously updated database is required.
One of the most important subspecific ranks applied to special forms of F. oxysporum are physiological pathotypes or races. This classification system is of great importance to plant breeders, especially for resistance breeding. Traditionally, race demarcation is based on cultivar specificity linked to specific resistance genes of the plant host cultivar (Armstrong & Armstrong 1981, Kistler 1997, Baayen et al. 2000, Roebroeck 2000, Fourie et al. 2011, Epstein et al. 2017). However, race designation has been inconsistent in the past (Gerlagh & Blok 1988, Correll 1991, Kistler 1997, Fourie et al. 2011) with several different nomenclatural systems being applied (Gabe 1975, Risser et al. 1976, Armstrong & Armstrong 1981) to further cause confusion (Kistler 1997). With advances in molecular technology, identification of races has been simplified using sequence-characterised amplified region (SCAR) primers (Lievens et al 2008, Epstein et al. 2017, Gilardi et al. 2017). However, time consuming and laborious pathogenicity tests are still needed to identify new emerging races and to test whether newly developed plant cultivars are resistant to known races (Epstein et al. 2017, Gilardi et al. 2017).
The use of vegetative compatibility (also known as heterokaryon compatibility) has formed an integral part of subspecific classification of F. oxysporum special forms and non-pathogenic strains. Formation of a stable heterokaryon between two auxotrophic nutritional mutants is regulated by several vic or het incompatibility loci (Correll 1991, Leslie 1993) indicating that the strains are homogenic at these loci (Correll 1991) and considered to be part of the same VCG. Therefore, classification using vegetative compatibility is based on genetic similarity at specific loci and not pathogenicity, providing a crude marker for population genetic studies (Correll 1991, Gordon & Martyn 1997, Leslie 1993, Leslie & Summerell 2006). Puhalla (1985), utilizing nit mutants, was the first to identify VCGs in F. oxysporum and characterised 16 VCGs in a collection of 21 F. oxysporum strains. The numbering system applied by Puhalla (1985), which is still used today, consists of a three-digit numerical code indicating the special form followed by digit(s) indicating the VCG (Katan 1999, Katan & Di Primo 1999). Conventional VCG characterisation is a relatively objective, time consuming and laborious assay only indicating genetic similarity and not genetic difference (Kistler 1997). Therefore, several PCR-based detection methods have been developed to identify economically important VCGs as diagnostic tool (Fernandez et al. 1998, Pasquali et al. 2004a, c, Lievens et al. 2008), e.g., F. oxysporum f. sp. cubense TR4 VCG01213 (Dita et al. 2010).
Until recently, limited knowledge on the genetic premise for host specificity in F. oxysporum was available (Gordon & Martyn 1997, Kistler 1997, Baayen et al. 2000). However, the discovery of a lineage-specific chromosome (or transposable/effector/accessory chromosome) in F. oxysporum f. sp. lycopersici by Ma et al. (2010), in which the host specific virulence genes lie (Van der Does et al. 2008, Takken & Rep 2010, Ma et al. 2013), has provided a new view into the evolution of pathogenicity in F. oxysporum. In vitro transfer of these accessory chromosomes into non-pathogenic F. oxysporum strains has converted the latter strains into host-specific pathogens, providing evidence that host-specific pathogenicity could be acquired through horizontal transfer of accessory chromosomes (Takken & Rep 2010, Ma et al. 2010, 2013, Van Dam et al. 2016, Van Dam & Rep 2017). Therefore, the special form name can be linked to the accessory chromosome whereas race demarcation can be linked to the specific virulence genes carried on these accessory chromosomes.
The genetic and functional mechanisms of the infection process in plants of various special forms of F. oxysporum has been well documented (Di Pietro et al. 2003, Ma et al. 2013, Upasani et al. 2016, Gordon 2017). However, these same mechanisms are still poorly understood in human and animal infections (O’Donnell et al. 2004, Guarro 2013, Van Diepeningen et al. 2015). Fusarium oxysporum has been linked to fungal keratitis (Hemo et al. 1989, Chang et al. 2006) and dermatitis (Guarro & Gene 1995, Romano et al. 1998, Ninet et al. 2005, Cutuli et al. 2015, Van Diepeningen et al. 2015), and has been isolated from contaminated hospital water systems (Steinberg et al. 2015, Edel-Hermann et al. 2016) and medical equipment (Barton et al. 2016, Carlesse et al. 2017) posing a serious threat to immunocompromised patients. Several recent reports also indicate that F. oxysporum is able to infect immunocompetent patients (Jiang et al. 2016, Khetan et al. 2018). In general, fusariosis is difficult to treat as Fusarium species display a remarkable resistance to antifungal agents (Guarro 2013, Al-Hatmi et al. 2018). However, some antimycotics are known to be effective against F. oxysporum related fusariosis (Al-Hatmi et al. 2018). Recently, both mycotoxins beauvericin and fusaric acid, produced by F. oxysporum strains that can infect tomato, have been shown to be important virulence determinants to infect immunosuppressed mice (López-Berges et al. 2013, López-Díaz et al. 2018).
Strains of F. oxysporum are known to produce a cocktail of polyketide secondary metabolites, some with unknown function and toxicities (Marasas et al. 1984, Mirocha et al. 1989, Bell et al. 2003, Desjardins 2006, Manici et al. 2017). Some of the better-known toxins produced by F. oxysporum include beauvericin (Marasas et al. 1984, Logrieco et al. 1998, López-Berges et al. 2013), fusaric acid (Marasas et al. 1984, López-Díaz et al. 2018) and fumonisins (Rheeder et al. 2002) to name a few. Mycotoxicological studies on F. oxysporum has thus far only focused on a strain to strain basis and therefore no link has yet been established between special form and/or race and mycotoxin production capabilities.
In light of the complicated and sometimes confusing classification systems applied to F. oxysporum taxonomy and nomenclature, the question has risen whether F. oxysporum truly represent a species (Kistler 1997). Given that F. oxysporum is a common, widespread, soil-borne fungus, with a global distribution and high economic importance, this question requires urgent attention. Therefore, to advance and stabilize the taxonomic and nomenclatural position of F. oxysporum and allow naming of the multiple cryptic species recognised in this species complex, Fusarium isolates were collected from the type locality in Berlin, Germany, and the type substrate, Solanum tuberosum. Using molecular phylogenetic and morphological tools, an epitype is designated for F. oxysporum in the present study based on these collections.
MATERIALS AND METHODS
Isolates
Tubers of S. tuberosum (potato), displaying symptoms of dry rot, were collected from several vegetable gardens in Berlin, Germany. Potato tubers were placed individually in paper bags, stored at 4 °C until transported to the laboratory for further processing. After surface-sterilisation of the potato tubers using a 10 % (v/v) sodium hypochlorite solution, pieces of symptomatic tissue were removed from the leading edges of the rot lesions and plated onto 2 % (w/v) potato dextrose agar (PDA) amended with 100 μg/mL penicillin and 100 μg/mL streptomycin, and peptone pentachloronitrobenzene agar (PCNB; Nash & Snyder 1962) and incubated at 25 °C in the dark. Axenic cultures were prepared on PDA from characteristic Fusarium colonies. Additional strains, previously identified as F. oxysporum, were obtained from the culture collection (CBS) of the Westerdijk Fungal Biodiversity Institute (WFBI), Utrecht, the Netherlands, and the working collection of Pedro W. Crous (CPC) housed at WFBI (Table 2).
Table 2.
Details of Fusarium strains included in the phylogenetic analyses.
| Species | Culture accession1 | Host/substrate | Special form | Origin | GenBank accession | ||||
|---|---|---|---|---|---|---|---|---|---|
| cmdA | IGS | rpb2 | tef1 | tub2 | |||||
| Fusarium callistephi | CBS 187.53T | Callistephus chinensis | callistephi | The Netherlands | MH484693 | MH484784 | MH484875 | MH484966 | MH485057 |
| CBS 115423 | Agathosma betulina | South Africa | MH484723 | MH484814 | MH484905 | MH484996 | MH485087 | ||
| F. carminascens | CBS 144739 = CPC 25792 | Zea mays | South Africa | MH484752 | MH484843 | MH484934 | MH485025 | MH485116 | |
| CBS 144740 = CPC 25793 | Z. mays | South Africa | MH484753 | MH484844 | MH484935 | MH485026 | MH485117 | ||
| CBS 144741 = CPC 25795 | Z. mays | South Africa | MH484754 | MH484845 | MH484936 | MH485027 | MH485118 | ||
| CBS 144738 = CPC 25800T | Z. mays | South Africa | MH484755 | MH484846 | MH484937 | MH485028 | MH485119 | ||
| F. contaminatum | CBS 111552 | Pasteurized fruit juice | The Netherlands | MH484718 | MH484809 | MH484900 | MH484991 | MH485082 | |
| CBS 114899T | Pasteurized chocolate milk | Germany | MH484719 | MH484810 | MH484901 | MH484992 | MH485083 | ||
| CBS 117461 | Tetra pack with milky nutrition | The Netherlands | MH484729 | MH484820 | MH484911 | MH485002 | MH485093 | ||
| F. cugenangense | CBS 620.72 = DSM 11271 = NRRL 36520 | Crocus sp. | gladioli | Germany | MH484697 | MH484788 | MH484879 | MH484970 | MH485061 |
| CBS 130304 = BBA 69050 = NRRL 25433 | Gossypium barbadense | vasinfectum | China | MH484739 | MH484830 | MH484921 | MH485012 | MH485103 | |
| CBS 130308 = ATCC 26225 = NRRL 25387 | Human toe nail | New Zealand | MH484738 | MH484829 | MH484920 | MH485011 | MH485102 | ||
| CBS 131393 | Vicia faba | Australia | MH484746 | MH484837 | MH484928 | MH485019 | MH485110 | ||
| F. curvatum | CBS 247.61 = BBA 8398 = DSM 62308 = NRRL 22545 | Matthiola incana | matthiolae | Germany | MH484694 | MH484785 | MH484876 | MH484967 | MH485058 |
| CBS 238.94 = NRRL 26422 = PD 94/184T | Beaucarnia sp. | meniscoideum | The Netherlands | MH484711 | MH484802 | MH484893 | MH484984 | MH485075 | |
| CBS 141.95 = NRRL 36251 = PD 94/1518 | Hedera helix | The Netherlands | MH484712 | MH484803 | MH484894 | MH484985 | MH485076 | ||
| F. duoseptatum | CBS 102026 = NRRL 36115 | Musa sapientum cv. Pisang ambon | cubense | Malaysia | MH484714 | MH484805 | MH484896 | MH484987 | MH485078 |
| F. elaeidis | CBS 217.49 = NRRL 36358 | Elaeis sp. | elaeidis | Zaire | MH484688 | MH484779 | MH484870 | MH484961 | MH485052 |
| CBS 218.49 = NRRL 36359 | Elaeis sp. | elaeidis | Zaire | MH484689 | MH484780 | MH484871 | MH484962 | MH485053 | |
| CBS 255.52 = NRRL 36386 | Elaeis guineensis | elaeidis | Unknown | MH484692 | MH484783 | MH484874 | MH484965 | MH485056 | |
| F. fabacearum | CBS 144742 = CPC 25801 | Z. mays | South Africa | MH484756 | MH484847 | MH484938 | MH485029 | MH485120 | |
| CBS 144743 = CPC 25802T | Glycine max | South Africa | MH484757 | MH484848 | MH484939 | MH485030 | MH485121 | ||
| CBS 144744 = CPC 25803 | G. max | South Africa | MH484758 | MH484849 | MH484940 | MH485031 | MH485122 | ||
| F. foetens | CBS 120665 | Nicotiana tabacum | Iran | MH484736 | MH484827 | MH484918 | MH485009 | MH485100 | |
| F. glycines | CBS 176.33 = NRRL 36286 | Linum usitatissium | lini | Unknown | MH484686 | MH484777 | MH484868 | MH484959 | MH485050 |
| CBS 214.49 = NRRL 36356 | Unknown | Argentina | MH484687 | MH484778 | MH484869 | MH484960 | MH485051 | ||
| CBS 200.89 | Ocimum basilicum | basilici | Italy | MH484706 | MH484797 | MH484888 | MH484979 | MH485070 | |
| CBS 144745 = CPC 25804 | G. max | South Africa | MH484759 | MH484850 | MH484941 | MH485032 | MH485123 | ||
| CBS 144746 = CPC 25808T | G. max | South Africa | MH484760 | MH484851 | MH484942 | MH485033 | MH485124 | ||
| F. gossypinum | CBS 116611 | Gossypium hirsutum | vasinfectum | Ivory Coast | MH484725 | MH484816 | MH484907 | MH484998 | MH485089 |
| CBS 116612 | G. hirsutum | vasinfectum | Ivory Coast | MH484726 | MH484817 | MH484908 | MH484999 | MH485090 | |
| CBS 116613T | G. hirsutum | vasinfectum | Ivory Coast | MH484727 | MH484818 | MH484909 | MH485000 | MH485091 | |
| F. hoodiae | CBS 132474T | Hoodia gordonii | hoodiae | South Africa | MH484747 | MH484838 | MH484929 | MH485020 | MH485111 |
| CBS 132476 | H. gordonii | hoodiae | South Africa | MH484748 | MH484839 | MH484930 | MH485021 | MH485112 | |
| CBS 132477 | H. gordonii | hoodiae | South Africa | MH484749 | MH484840 | MH484931 | MH485022 | MH485113 | |
| F. languescens | CBS 645.78 = NRRL 36531T | Solanum lycopersicum | lycopersici | Morocco | MH484698 | MH484789 | MH484880 | MH484971 | MH485062 |
| CBS 646.78 = NRRL 36532 | S. lycopersicum | lycopersici | Morocco | MH484699 | MH484790 | MH484881 | MH484972 | MH485063 | |
| CBS 413.90 = ATCC 66046 = NRRL 36465 | S. lycopersicum | lycopersici | Israel | MH484708 | MH484799 | MH484890 | MH484981 | MH485072 | |
| CBS 300.91 = NRRL 36416 | S. lycopersicum | lycopersici | The Netherlands | MH484709 | MH484800 | MH484891 | MH484982 | MH485073 | |
| CBS 302.91 = NRRL 36419 | S. lycopersicum | lycopersici | The Netherlands | MH484710 | MH484801 | MH484892 | MH484983 | MH485074 | |
| CBS 872.95 = NRRL 36570 | S. lycopersicum | radicis-lycopersici | Unknown | MH484713 | MH484804 | MH484895 | MH484986 | MH485077 | |
| CBS 119796 = MRC 8437 | Z. mays | South Africa | MH484735 | MH484826 | MH484917 | MH485008 | MH485099 | ||
| F. libertatis | CBS 144748 = CPC 25782 | Aspalathus sp. | South Africa | MH484750 | MH484841 | MH484932 | MH485023 | MH485114 | |
| CBS 144747 = CPC 25788 | Aspalathus sp. | South Africa | MH484751 | MH484842 | MH484933 | MH485024 | MH485115 | ||
| CBS 144749 = CPC 28465T | Rock surface | South Africa | MH484762 | MH484853 | MH484944 | MH485035 | MH485126 | ||
| F. nirenbergiae | CBS 129.24 | Secale cereale | Unknown | MH484682 | MH484773 | MH484864 | MH484955 | MH485046 | |
| CBS 149.25 = NRRL 36261 | Musa sp. | cubense | Unknown | MH484683 | MH484774 | MH484865 | MH484956 | MH485047 | |
| CBS 181.32 = NRRL 36303 | S. tuberosum | USA | MH484685 | MH484776 | MH484867 | MH484958 | MH485049 | ||
| CBS 758.68 = NRRL 36546 | S. lycopersicum | lycopersici | The Netherlands | MH484695 | MH484786 | MH484877 | MH484968 | MH485059 | |
| CBS 744.79 = BBA 62355 = NRRL 22549 | Passiflora edulis | passiflorae | Brazil | MH484700 | MH484791 | MH484882 | MH484973 | MH485064 | |
| CBS 127.81 = BBA 63924 = NRRL 36229 | Chrysanthemum sp. | chrysanthemi | USA | MH484701 | MH484792 | MH484883 | MH484974 | MH485065 | |
| CBS 129.81 = BBA 63926 = NRRL 22539 | Chrysanthemum sp. | chrysanthemi | USA | MH484703 | MH484794 | MH484885 | MH484976 | MH485067 | |
| CBS 196.87 = NRRL 26219 | Bouvardia longiflora | bouvardiae | Italy | MH484704 | MH484795 | MH484886 | MH484977 | MH485068 | |
| CBS 840.88T | Dianthus caryophyllus | dianthi | The Netherlands | MH484705 | MH484796 | MH484887 | MH484978 | MH485069 | |
| CBS 115416 = CPC 5307 | Agathosma betulina | South Africa | MH484720 | MH484811 | MH484902 | MH484993 | MH485084 | ||
| CBS 115417 = CPC 5306 | A. betulina | South Africa | MH484721 | MH484812 | MH484903 | MH484994 | MH485085 | ||
| CBS 115419 = CPC 5308 | A. betulina | South Africa | MH484722 | MH484813 | MH484904 | MH484995 | MH485086 | ||
| CBS 115424 = CPC 5312 | A. betulina | South Africa | MH484724 | MH484815 | MH484906 | MH484997 | MH485088 | ||
| CBS 123062 = GJS 91-17 | Tulip roots | USA | MH484737 | MH484828 | MH484919 | MH485010 | MH485101 | ||
| CBS 130300 = NRRL 26368 | Amputated human toe | USA | MH484743 | MH484834 | MH484925 | MH485016 | MH485107 | ||
| CBS 130301 = NRRL 26374 | Human leg ulcer | USA | MH484744 | MH484835 | MH484926 | MH485017 | MH485108 | ||
| CBS 130303 | S. lycopersicum | radicis-lycopersici | USA | MH484741 | MH484832 | MH484923 | MH485014 | MH485105 | |
| CPC 30807 | South Africa | MH484768 | MH484859 | MH484950 | MH485041 | MH485132 | |||
| F. odoratissimum | CBS 794.70 = BBA 11103 = NRRL 22550 | Albizzia julibrissin | perniciosum | Iran | MH484696 | MH484787 | MH484878 | MH484969 | MH485060 |
| CBS 102030 | M. sapientum cv. Pisang mas | cubense | Malaysia | MH484716 | MH484807 | MH484898 | MH484989 | MH485080 | |
| CBS 130310 = NRRL 25603 | Musa sp. | cubense | Australia | MH484740 | MH484831 | MH484922 | MH485013 | MH485104 | |
| F. oxysporum | CBS 221.49 = IHEM 4508 = NRRL 22546 | Camellia sinensis | medicaginis | South East Asia | MH484690 | MH484781 | MH484872 | MH484963 | MH485054 |
| CBS 144134ET | S. tuberosum | Germany | MH484771 | MH484862 | MH484953 | MH485044 | MH485135 | ||
| CBS 144135 | S. tuberosum | Germany | MH484772 | MH484863 | MH484954 | MH485045 | MH485136 | ||
| CPC 25822 | Protea sp. | South Africa | MH484761 | MH484852 | MH484943 | MH485034 | MH485125 | ||
| F. pharetrum | CBS 144750 = CPC 30822 | Aliodendron dichotomum | South Africa | MH484769 | MH484860 | MH484951 | MH485042 | MH485133 | |
| CBS 144751 = CPC 30824T | A. dichotomum | South Africa | MH484770 | MH484861 | MH484952 | MH485043 | MH485134 | ||
| F. trachichlamydosporum | CBS 102028 = NRRL 36117 | M. sapientum cv. Pisang awak legor | cubense | Malaysia | MH484715 | MH484806 | MH484897 | MH484988 | MH485079 |
| F. triseptatum | CBS 258.50 = NRRL 36389T | Ipomoea batatas | batatas | USA | MH484691 | MH484782 | MH484873 | MH484964 | MH485055 |
| CBS 116619 | G. hirsutum | vasinfectum | Ivory Coast | MH484728 | MH484819 | MH484910 | MH485001 | MH485092 | |
| CBS 119665 | Sago starch | Papua New Guinea | MH484734 | MH484825 | MH484916 | MH485007 | MH485098 | ||
| CBS 130302 = NRRL 26360 = FRC 755 | Human eye | USA | MH484742 | MH484833 | MH484924 | MH485015 | MH485106 | ||
| F. udum | CBS 177.31 | Digitaria eriantha | South Africa | MH484684 | MH484775 | MH484866 | MH484957 | MH485048 | |
| F. veterinarium | CBS 109898 = NRRL 36153T | Shark peritoneum | The Netherlands | MH484717 | MH484808 | MH484899 | MH484990 | MH485081 | |
| CBS 117787 | Swab sample near filling apparatus | The Netherlands | MH484730 | MH484821 | MH484912 | MH485003 | MH485094 | ||
| CBS 117790 | Swab sample near filling apparatus | The Netherlands | MH484731 | MH484822 | MH484913 | MH485004 | MH485095 | ||
| CBS 117791 | Pasteurized milk-based product | The Netherlands | MH484732 | MH484823 | MH484914 | MH485005 | MH485096 | ||
| CBS 117792 | Pasteurized milk-based product | The Netherlands | MH484733 | MH484824 | MH484915 | MH485006 | MH485097 | ||
| NRRL 54984 | Mouse mucosa | USA | MH484763 | MH484854 | MH484945 | MH485036 | MH485127 | ||
| NRRL 54996 | Little blue penguin foot | USA | MH484764 | MH484855 | MH484946 | MH485037 | MH485128 | ||
| NRRL 62542 | Unknown animal faeces | USA | MH484765 | MH484856 | MH484947 | MH485038 | MH485129 | ||
| NRRL 62545 | Endoscope of veterinary clinic | USA | MH484766 | MH484857 | MH484948 | MH485039 | MH485130 | ||
| NRRL 62547 | Canine stomach | USA | MH484767 | MH484858 | MH484949 | MH485040 | MH485131 | ||
| Fusarium sp. | CBS 128.81 = BBA 63925 = NRRL 36233 | Chrysanthemum sp. | chrysanthemi | USA | MH484702 | MH484793 | MH484884 | MH484975 | MH485066 |
| CBS 680.89 = NRRL 26221 | Cucumis sativus | cucurbitacearum | The Netherlands | MH484707 | MH484798 | MH484889 | MH484980 | MH485071 | |
| CBS 130323 | Human nail | Australia | MH484745 | MH484836 | MH484927 | MH485018 | MH485109 | ||
1ATCC: American Type Culture Collection, USA; BBA: Biologische Bundesanstalt für Land-und Forstwirtschaft, Berlin-Dahlem, Germany; CBS: Westerdijk Fungal Biodiverity Institute (WIFB), Utrecht, The Netherlands; CPC: Collection of P.W. Crous; DSM: Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany; FRC: Fusarium Research Center, Penn State University, Pennsylvania; GJS: Collection of Gary J. Samuels; IHEM: Institute of Hygiene and Epidemiology-Mycology Laboratory, Brussels, Belgium; MRC: National Research Institute for Nutritional Diseases, Tygerberg, South Africa; NRRL: Agricultural Research Service Culture Collection, USA; PD: Collection of the Dutch National Plant Protection Organization, Wageningen, The Netherlands. T Ex-type culture; ETEpitype.
DNA isolation, PCR and sequencing
Total genomic DNA was extracted from isolates grown for 7 d on PDA at 24 °C using a 12/12 h photoperiod using the Wizard® Genomic DNA purification Kit (Promega Corporation, Madison, WI, USA), according to the manufacturer’s instructions. Partial gene sequences were determined for the β-tubulin (tub2), calmodulin (cmdA), the intergenic spacer region of the rDNA (IGS), RNA polymerase II second largest subunit (rpb2) and translation elongation factor 1-alpha (tef1), using PCR protocols described elsewhere (O’Donnell et al. 1998, 2007, 2009, 2010, Lombard et al. 2015). Primer pairs T1/CYLTUB1R (O’Donnell & Cigelnik 1997, Crous et al. 2004) for tub2, Cal228F/CAL2Rd (Carbone & Kohn 1999, Groenewald et al. 2013) for cmdA, iNL11/iCNS1 and the internal sequencing primers NLa/CNSa (O’Donnell et al. 2009) for IGS, 5f2/7cr (Liu et al. 1999, Sung et al. 2007) for rpb2, and EF1/EF2 (O’Donnell et al. 1998) for tef1, were used for amplifications of the respective gene regions. Integrity of the sequences was ensured by sequencing the amplicons in both directions using the same primer pairs as were used for amplification. Consensus sequences for each locus were assembled in MEGA v. 7 (Kumar et al. 2016), with the exception of the IGS locus, which was assembled in Geneious R11 (Kearse et al. 2012). All sequences generated in this study were deposited in GenBank (Table 1).
Phylogenetic analyses
Sequences of the individual loci were aligned using MAFFT v. 7.110 (Katoh et al. 2017) and manually corrected where necessary. The individual gene datasets were assessed for incongruency prior to concatenation using a 70 % reciprocal bootstrap criterion (Mason-Gamer & Kellogg 1996). Three independent phylogenetic algorithms, Maximum Parsimony (MP), Maximum Likelihood (ML) and Bayesian inference (BI), were employed for phylogenetic analyses. Phylogenetic analyses were conducted for the individual loci and then as a multilocus sequence dataset that included the cmdA, rpb2, tef1 and tub2 sequences.
For BI and ML, the best evolutionary models for each locus were determined using MrModeltest (Nylander 2004) and incorporated into the analyses. MrBayes v. 3.2.1 (Ronquist & Huelsenbeck 2003) was used for BI to generate phylogenetic trees under optimal criteria for each locus. A Markov Chain Monte Carlo (MCMC) algorithm of four chains was initiated in parallel from a random tree topology with the heating parameter set at 0.3. The MCMC analysis lasted until the average standard deviation of split frequencies was below 0.01 with trees saved every 1000 generations. The first 25 % of saved trees were discarded as the ‘burn-in’ phase and posterior probabilities (PP) were determined from the remaining trees.
The ML analyses were performed using RAxML v. 8.2.9 (randomised accelerated (sic) maximum likelihood for high performance computing; Stamatakis 2014) through the CIPRES website (http://www.phylo.org) to obtain another measure of branch support. The robustness of the analysis was evaluated by bootstrap support (BS) with the number of bootstrap replicates automatically determined by the software. For MP, analyses were done using PAUP (Phylogenetic Analysis Using Parsimony, v. 4.0b10; Swofford 2003) with phylogenetic relationships estimated by heuristic searches with 1 000 random addition sequences. Tree-bisection-reconnection was used, with branch swapping option set on ‘best trees’ only. All characters were weighted equally and alignment gaps treated as fifth state. Measures calculated for parsimony included tree length (TL), consistency index (CI), retention index (RI) and rescaled consistence index (RC). Bootstrap (BS) analyses (Hillis & Bull 1993) were based on 1 000 replications. Alignments and phylogenetic trees derived from this study were uploaded to TreeBASE (www.treebase.org).
Genealogical concordance phylogenetic species recognition (GCPSR)
In order to establish the recombination levels between the newly proposed species in this study and their closest phylogenetic relatives, pairwise homoplasy index (PHI) analyses were done on the respective concatenated multilocus datasets (Bruen et al. 2006). The analyses were conducted as described by Quaedvlieg et al. (2014) using SplitsTree v. 4.14.4 (Huson & Bryant 2006). Therefore, a PHI value below 0.05 (ϕW < 0.05) would indicate the presence of significant recombination in the dataset. Split graphs were constructed for visualization of the relationships between closely related species.
Morphological characterisation
All isolates were characterised following the protocols described by Leslie & Summerell (2006) using potato dextrose agar (PDA; recipe in Crous et al. 2009), synthetic nutrient-poor agar (SNA; Nirenberg 1976) and carnation leaf agar (CLA; Fisher et al. 1982). Colony morphology, pigmentation, odour and growth rates were evaluated on PDA after 3 and 7 d at 24 °C with a 12/12 h cool fluorescent light/dark cycle as described by Sandoval-Denis et al. (2018) and using the colour charts of Rayner (1970). Micromorphological characters were examined using water as mounting medium on a Zeiss Axioskop 2 plus with Differential Interference Contrast (DIC) optics and a Nikon AZ100 stereomicroscope both fitted with Nikon DS-Ri2 high definition colour digital cameras to photo-document fungal structures. Measurements were taken using the Nikon software NIS-elements D v. 4.50 and the 95 % confidence levels were determined for the conidial measurements with extremes given in parentheses. For all other fungal structures examined, only the extremes are presented. To facilitate the comparison of relevant micro- and macroconidial features, composite photo plates were assembled from separate photographs using PhotoShop CSS.
RESULTS
Isolates
A total of 23 fusarium-like isolates were obtained from the symptomatic tissues of the potato tubers. Of these, six isolates displayed typical F. oxysporum-like phenotypes, of which two (CBS 144134 and CBS 144135) were selected for further study.
Phylogenetic analyses
Approximately 500–650 bases were determined for cmdA, tef1 and tub2, 880 bases for rpb2 and 2 650 bases for IGS. Sequence comparisons of the IGS, rpb2 and tef1 gene regions generated in this study, against those in the Fusarium-ID (http://isolate.fusariumdb.org/blast.php) and Fusarium-MLST (http://www.westerdijkinstitute.nl/fusarium/) databases revealed that all isolates included in this study belonged to the FOSC. The congruency analysis revealed no conflict between the cmdA, rpb2, tef1 and tub2 sequence datasets and were therefore combined. However, the IGS sequence dataset revealed major conflict with several included taxa resolving into single lineages due to the large number of ambiguous regions in this gene region. Therefore, the IGS sequences were excluded from further analyses.
For the BI and ML analyses, a K80 model for cmdA, an HKY+ G+I model for rpb2, an HKY+G for tef1 and SYM+I+G model for tub2 were selected and incorporated into the analyses. The ML tree topology confirmed the tree topologies obtained from the BI and MP analyses, and therefore, only the ML tree is presented.
The combined four loci sequence dataset included 89 ingroup taxa with F. foetens (CBS 120665) and F. udum (CBS 177.31) as outgroup taxa. The dataset consisted of 2 679 characters including gaps. Of these characters, 2 291 were constant, 211 parsimony-uninformative and 177 parsimony-informative. The BI lasted for 1.2 M generations, and the consensus tree and posterior probabilities (PP) were calculated from 8 814 trees left after 2 937 were discarded as the ‘burn-in’ phase. The MP analysis yielded 1 000 trees (TL = 574; CI = 0.747; RI = 0.858; RC = 0.641) and a single best ML tree with -InL = 7353.014512 (Fig. 1).
Fig. 1.
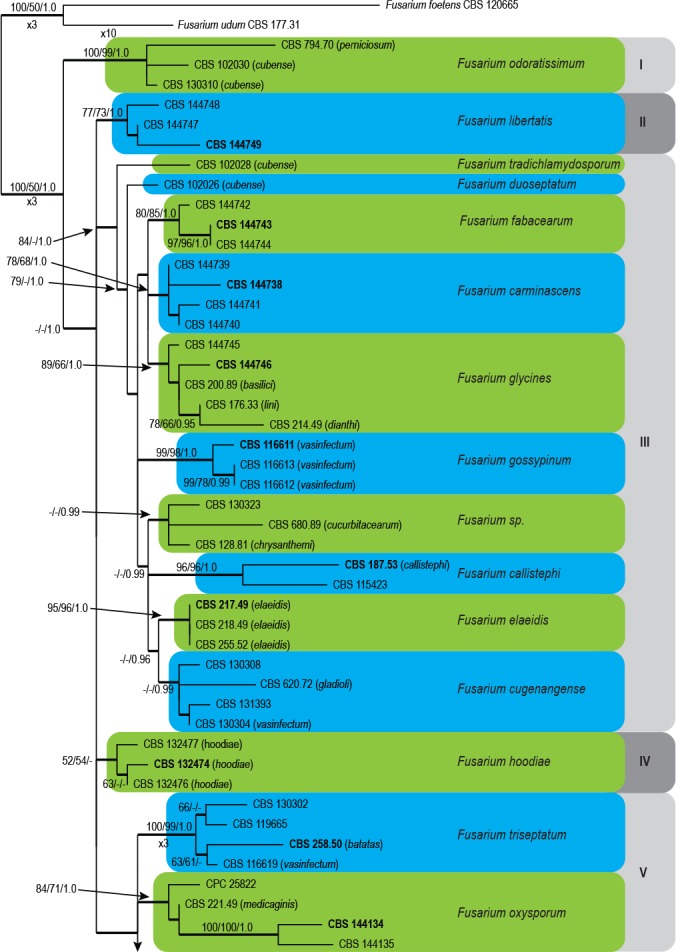
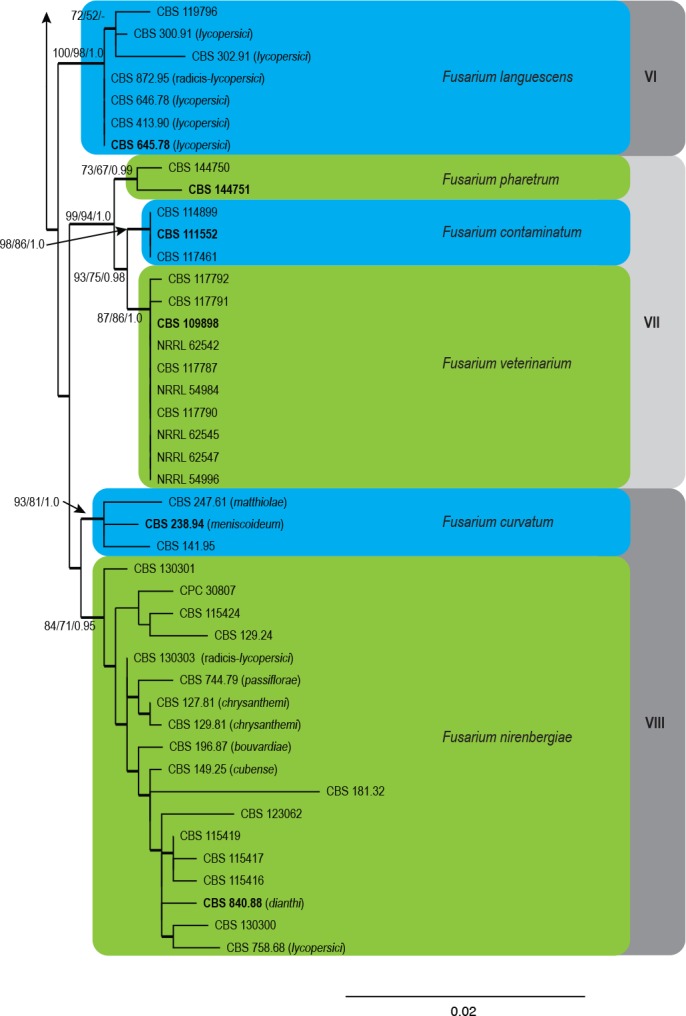
The ML consensus tree inferred from the combined cmdA, rpb2, tef1 and tub2 sequence alignment. Thickened branches indicate branches present in the ML, MP and Bayesian consensus trees. Support values (ML & MP bootstrap and posterior probability values) are indicated at the branches. The scale bar indicates 0.02 expected changes per site. Clade numbers are provided on the right of the tree and these are used for reference in the treatment of the species. The tree is rooted to F. foetens (CBS 120665) and F. udum (CBS 177.31). Epi- and ex-type strains are indicated in bold.
In the phylogenetic tree (Fig. 1) the ingroup taxa resolved into eight clades (I–VIII). Of these, Clades I, II, IV and VI represent single well- (ML & MP-BS ≥ 75–95 %; PP ≥ 0.95–0.98) to highly (ML & MP-BS ≥ 96 %; PP ≥ 0.99–1.0) supported clades, whereas Clades III, V, VII and VIII displayed substantial substructure. Clade III included eight well- to highly supported subclades as well as two single lineages. Sequence comparisons of the rpb2 and tef1 sequences with those generated by Maryani et al. (2019) revealed that both single lineages represented F. duoseptatum (CBS 102026) and F. tradichlamydosporum (CBS 102028), respectively. Similarly, the subclade that include isolates CBS 620.72, CBS 130304, CBS 130308 and CBS 131393 represent F. cugenangense. Both Clades V and VIII resolved two subclades in each, and Clade VII included three subclades. The phylogenetic relationships between Clades I–VIII and their underlying subclades are further discussed in the notes in the Taxonomy section.
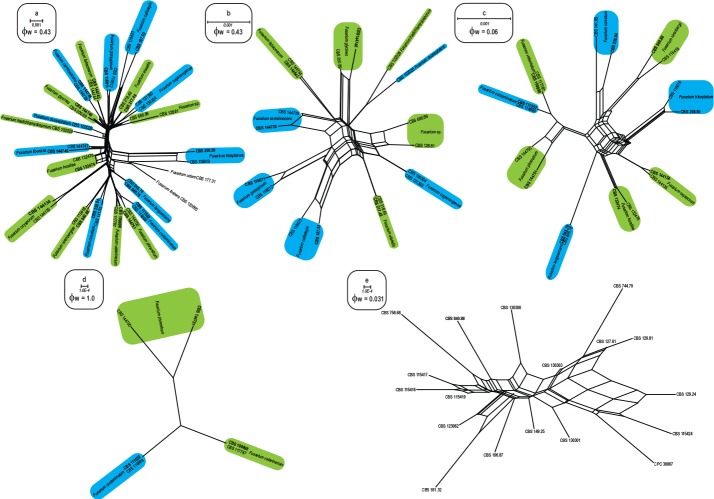
The PHI tests revealed that no evidence of recombination (ϕW = 0.43; Fig. 2a) was detected between each Clade (I–VIII) and their underlining subclades. Similarly, the genealogical exclusivity of the subclades in Clades III (ϕW = 0.43; Fig. 2b) and VII (ϕW = 1.0; Fig. 2d), as well as between Clades IV–VIII (ϕW = 0.06; Fig. 2c) was also confirmed. The basal subclade in Clade VIII (ϕW = 0.031; Fig. 2c), however, showed significant evidence for recombination among all isolates included.
Fig. 2.
Splitgraphs showing the results of the pairwise homoplasy index (PHI) test of newly described taxa using both LogDet transformation and splits decomposition. PHI test results (ϕW) < 0.05 indicate significant recombination within the dataset. a. Representatives of all phylogenetic species resolved in this study; b. phylogenetic species in Clade III; c. phylogenetic species in Clades IV–VIII; d. phylogenetic species in Clade VII; e. isolates representing F. nirenbergiae.
Taxonomy
In this section we provide a new (emended) description of F. oxysporum and designate an epitype for this species. The following species are also recognised as new within the FOSC, based on phylogenetic inference and morphological comparisons. Isolates CBS 128.81, CBS 680.89 and CBS 130323 in Clade III are not treated further as these were sterile.
Fusarium callistephi L. Lombard & Crous, sp. nov. — MycoBank MB826833; Fig. 3
Fig. 3.
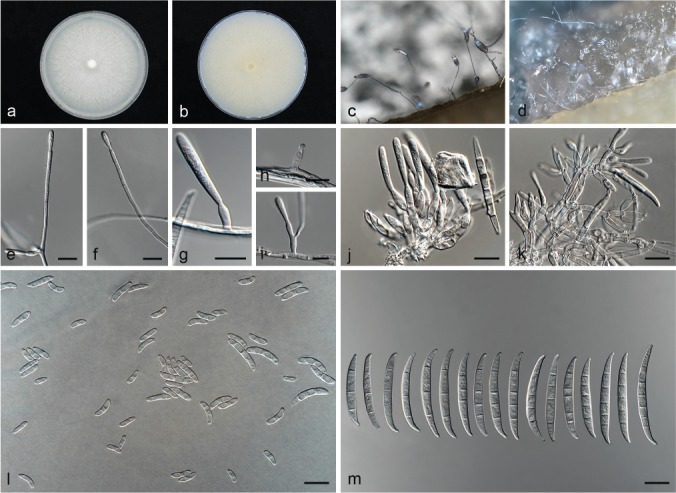
Fusarium callistephi (ex-type culture CBS 187.53). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c. conidiophores on surface of carnation leaf; d. sporodochia on carnation leaves; e–i. conidiophores and phialides on aerial mycelium; j–k. sporodochia and sporodochial conidiophores; l. aerial conidia (microconidia); m. sporodochial conidia (macroconidia). — Scale bars: e–m = 10 μm.
Etymology. Name refers to the plant genus Callistephus from which this fungus was isolated.
Typus. Netherlands, Oostenbrink, from Callistephus chinensis, 28 Feb. 1953, collector unknown (holotype CBS H-23608 designated here, culture ex-type CBS 187.53).
Conidiophores carried on the aerial mycelium 60–110 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 2–23 × 3–4 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (6–)7–11(–14) × 2–3 μm (av. 9 × 3 μm); 1-septate conidia: (13–)14–18(–20) × 2–4 μm (av. 16 × 3 μm). Sporodochia pale luteous to pale rosy vinaceous, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 4–7 × 2–4 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 9–13 × 3–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, 3–4(–5)-septate, hyaline, smooth- and thin-walled; 3-septate conidia: (28–)33–39(–40) × 3–5 μm (av. 36 × 4 μm); 4-septate conidia: (30–)35–41(–42) × 3–5 μm (av. 38 × 4 μm); 5-septate conidia: 36–44(–47) × 4–5 μm (av. 40 × 5 μm). Chlamydospores not observed.
Culture characteristics — Colonies on PDA with an average radial growth rate of 2.9–4.2 mm/d at 24 °C. Colony surface white to pale vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse colourless, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, lacking chlamydospores, aerial mycelium sparse with moderate sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant pale luteous to pale rosy vinaceous sporodochia forming on the carnation leaves.
Additional material examined. South Africa, Western Cape Province, Piketberg, from Agathosma betulina, 2001, K. Lubbe, CBS 115423 = CPC 5311.
Notes — Fusarium callistephi formed a highly-supported subclade in Clade III, closely related to F. cugenangense, F. elaeidis and the untreated Fusarium clade. This species (conidia 3–4(–5)-septate) can be distinguished from F. cugenangense (conidia 3–6-septate; Maryani et al. 2019) and F. elaeidis ((1–)3–5-septate) based on septation of their macroconidia. Additionally, F. cugenangense produces up to 3-septate microconidia, a feature not seen in either F. callistephi or F. elaeidis. Fusarium elaeidis readily formed polyphialidic conidiogenous cells on the aerial mycelium, not seen in F. callistephi.
Fusarium carminascens L. Lombard, Crous & Lampr., sp. nov. — MycoBank MB826835; Fig. 4
Fig. 4.
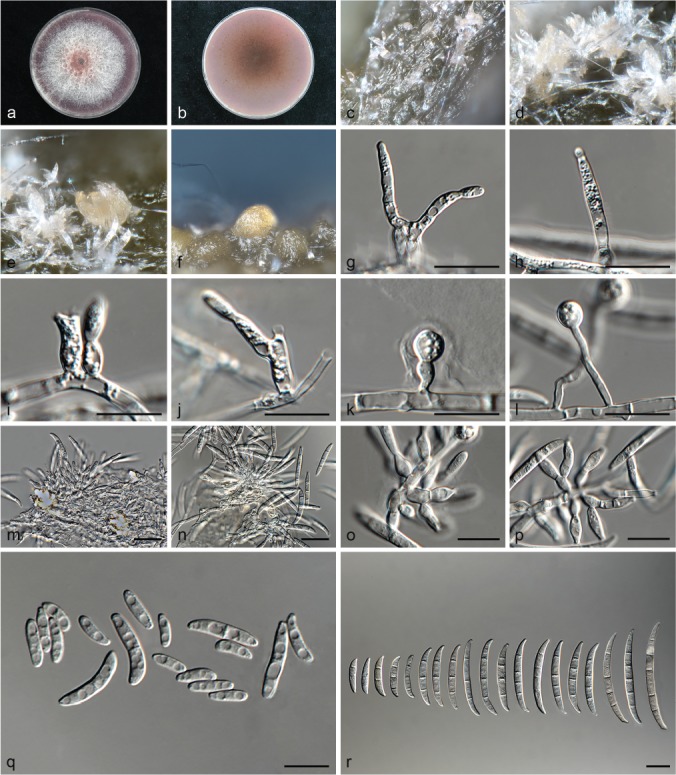
Fusarium carminascens (ex-type culture CBS 144738). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. sporodochia on carnation leaves; g–j. conidiophores and phialides on aerial mycelium; g–h. monophialides; i–j. polyphialides; k–l. chlamydospores; m–p. sporodochia and sporodochial conidiophores; o–p. phialides of sporodochial conidiophores; q. aerial conidia (microconidia); r. sporodochial conidia (macroconidia). — Scale bars: g–r = 10 μm.
Etymology. Name refers to the almost carmine exudates this fungus produces in its aerial mycelium when grown on PDA.
Typus. South Africa, KwaZulu-Natal Province, from Zea mays, 2008, S.C. Lamprecht (holotype CBS H-23609 designated here, culture ex-type CBS 144738 = CPC 25800).
Conidiophores carried on the aerial mycelium 35–55 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily phialides, often reduced to single phialides; aerial phialides mono- and polyphialidic, subulate to subcylindrical, smooth- and thin-walled, 8–18 × 3–4 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (5–)7–11(–12) × 2–3(–4) μm (av. 9 × 3 μm); 1-septate conidia: (12–)13–15(–18) × 2–4 μm (av. 14 × 3 μm). Sporodochia bright orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 4–9 × 2–4 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 5–13 × 2–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (2–)3–4(–5)-septate, hyaline, smooth- and thin-walled; 2-septate conidia: 16–19 × 3–4 μm (av. 18 × 3 μm); 3-septate conidia: (21–)26–36(–40) × 3–5 μm (av. 31 × 4 μm); 4-septate conidia: (31–)33–43(–44) × 4–5 μm (av. 38 × 4 μm); 5-septate conidia: 45–51 × 4 μm (av. 48 × 4 μm). Chlamydospores globose to subglobose, formed terminally, 4–8 μm diam.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.1–4.0 mm/d at 24 °C. Colony surface vinaceous purple to livid purple, floccose with abundant aerial mycelium which produce an almost carmine exudate; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse dark livid to livid purple, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, with abundant chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant bright orange sporodochia forming on the carnation leaves.
Additional materials examined. South Africa, KwaZulu-Natal Province, from Zea mays, 2008, S.C. Lamprecht, CBS 144739 = CPC 25792, CBS 144740 = CPC 25793, CBS 144741 = CPC 25795.
Notes — Fusarium carminascens formed a well-supported subclade in Clade III, closely related to F. fabacearum and F. glycines. This species produced an almost carmine coloured exudate in its aerial mycelium, a feature not observed in any of the other strains studied here. Furthermore, F. carminascens produces polyphialidic conidiogenous cells on its aerial mycelium, not observed in F. fabacearum or F. glycines.
Fusarium contaminatum L. Lombard & Crous, sp. nov. — Myco-Bank MB826836; Fig. 5
Fig. 5.
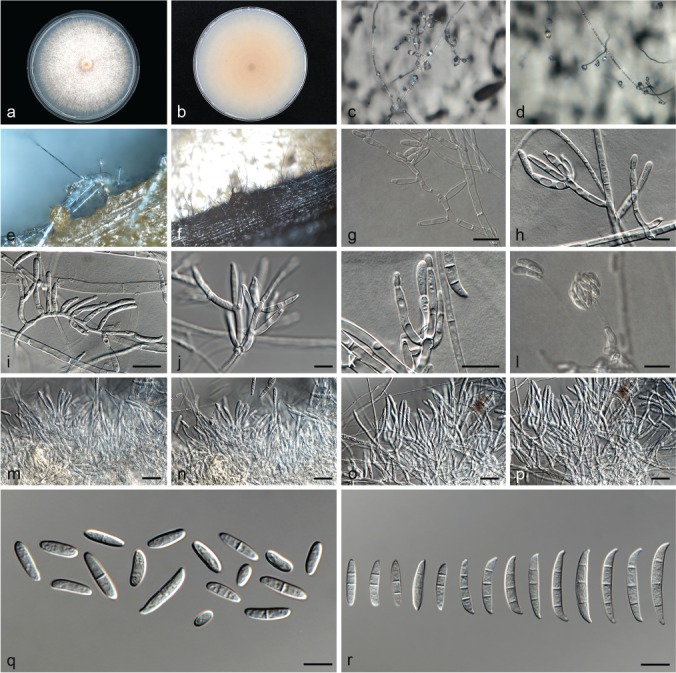
Fusarium contaminatum (ex-type culture CBS 114899). a–b. Colony on PDA; a. Surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. sporodochia on carnation leaves; g–k. conidiophores and phialides on aerial mycelium; l. false head carried on phialide on aerial mycelium; m–p. sporodochia and sporodochial conidiophores; q. aerial conidia (microconidia); r. sporodochial conidia (macroconidia). — Scale bars: g–l, q–r = 10 μm; m–p = 20 μm.
Etymology. Name refers to the fact that this fungus was isolated from contaminated food products.
Typus. Germany, Schluchtern, from pasteurized chocolate milk, Apr. 2004, J. Houbraken (holotype CBS H-23610 designated here, culture ex-type CBS 114899).
Conidiophores carried on the aerial mycelium 15–85 μm tall, unbranched or branched, bearing a single terminal or a whorl of 2–4 monophialides or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 7–22 × 2–5 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: 5–9(–11) × 2–4 μm (av. 7 × 3 μm); 1-septate conidia: (9–)10–14(–17) × 2–4 μm (av. 12 × 3 μm). Sporodochia bright orange, formed sparsely on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 7–13 × 4 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 4–9 × 2–3 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (2–)3-septate, hyaline, smooth- and thin-walled; 2-septate conidia: (14–)15–17 × 3–4 μm (av. 16 × 3 μm); 3-septate conidia: (18–)20–26(–28) × 3–5 μm (av. 23 × 4 μm). Chlamydospores not observed.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.1–4.5 mm/d at 24 °C. Colony surface white to pale vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse rosy vinaceous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, lacking chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant orange sporodochia forming on the carnation leaves.
Additional materials examined. Netherlands, from pasteurized fruit juice, date and collector unknown, CBS 111552; from tetra pack with milky nutrition, 2005, collector unknown, CBS 117461.
Notes — Fusarium contaminatum formed a highly-supported subclade in Clade VII, closely related to F. pharetrum and F. veterinarium. This species produces small, 2–3-septate macroconidia, whereas F. pharetrum produces much larger, 3(–4)-septate macroconidia and F. veterinarium produces slightly smaller, 1–(2–)3-septate macroconidia. None of these three species produced any chlamydospores on SNA.
Fusarium curvatum L. Lombard & Crous, sp. nov. — MycoBank MB826837; Fig. 6
Fig. 6.
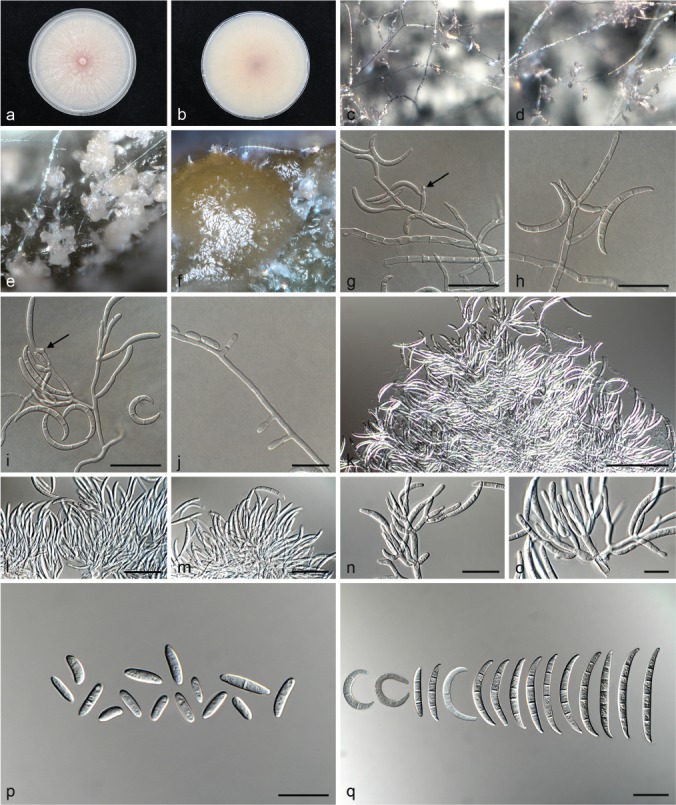
Fusarium curvatum (ex-type culture CBS 238.94). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. sporodochia on carnation leaves; g–i. conidiophores, monophialides and polyphialides (arrows) on aerial mycelium; j. phialidic pegs on aerial mycelium; k–o. sporodochia and sporodochial conidiophores; p. aerial conidia (microconidia); q. sporodochial conidia (macroconidia). — Scale bars: g–i, n = 20 μm; j, o–q = 10 μm, k–m = 50 μm.
Etymology. Name refers to the strongly curved sporodochial conidia produced by this fungus.
Typus. Netherlands, from Beaucarnia sp., 1994, J.W. Veenbaas-Rijks (holo-type CBS H-23611 designated here, culture ex-type CBS 238.94 = NRRL 26422 = PD 94/184).
Conidiophores carried on the aerial mycelium 25–56 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily phialides, often reduced to single phialides or as phialidic pegs; aerial phialides mono- and polyphialidic, subulate to subcylindrical, smooth- and thin-walled, 3–30 × 2–5 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (4–)5–9(–11) × 2–4 μm (av. 7 × 3 μm); 1-septate conidia: (10–)11–13 × 2–4 μm (av. 12 × 3 μm). Sporodochia orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 8–10 × 2–4 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 8–22 × 2–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, strongly curved or curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (2–)3–5-septate, hyaline, smooth- and thin-walled; 2-septate conidia: (15–)16–22(–23) × 3–4 μm (av. 19 × 3 μm); 3-septate conidia: (18–)27–39(–41) × 3–5 μm (av. 33 × 4 μm); 4-septate conidia: (34–)37–43(–46) × 3–5 μm (av. 40 × 4 μm); 5-septate conidia: (30–)38–46(–51) × 3–5 μm (av. 42 × 4 μm). Chlamydospores not observed.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.1–4.5 mm/d at 24 °C. Colony surface pale vinaceous to rosy vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse pale vinaceous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, lacking chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant orange sporodochia forming on the carnation leaves.
Additional materials examined. Germany, Berlin-Dahlem, from Matthiola incana, Feb. 1957, W. Gerlach, CBS 247.61 = BBA 8398 = DSM 62308 = NRRL 22545. – Netherlands, from Hedera helix, 1994, J.W. Veenbaas-Rijks, CBS 141.95 = NRRL 36251 = PD 94/1518.
Notes — Fusarium curvatum formed a highly-supported subclade in Clade VIII, closely related to F. nirenbergiae. This species produces strongly curved 3-septate macroconidia and aerial polyphialidic conidiogenous cells, distinguishing it from F. nirenbergiae. Additionally, F. curvatum failed to produce any chlamydospores on SNA, whereas F. nirenbergiae produced abundant chlamydospores.
Fusarium elaeidis L. Lombard & Crous, sp. nov. — MycoBank MB826838; Fig. 7
Fig. 7.
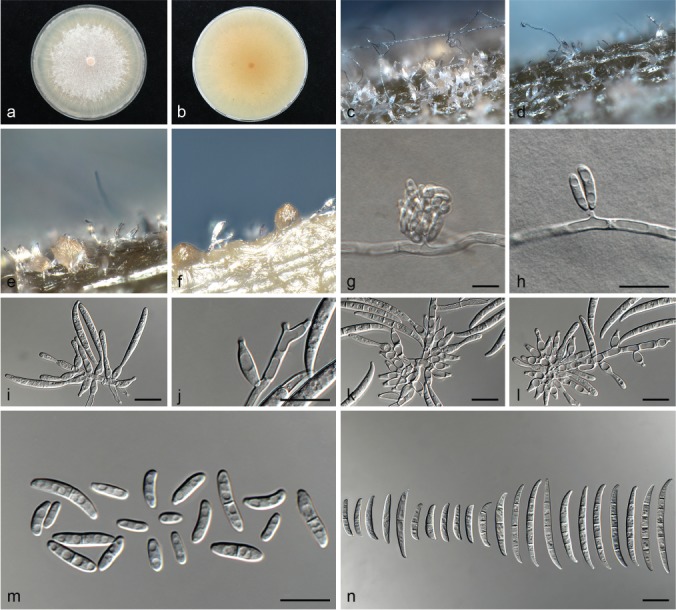
Fusarium elaeidis (ex-type culture CBS 217.49). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. sporodochia on carnation leaves; g. false head carried on a phialidic peg on aerial mycelium; h. phialidic peg; i–j. conidiophores and phialides on aerial mycelium; j. polyphialide; k–l. sporodochia and sporodochial conidiophores; m. aerial conidia (microconidia); n. sporodochial conidia (macroconidia). — Scale bars: g–n = 10 μm.
Etymology. Name refers to the host plant genus Elaeis, from which this fungus was first isolated.
Typus. Zaire, from Elaeis sp., 1949, T. Gogoi (holotype CBS H-23612 designated here, culture ex-type CBS 217.49 = NRRL 36358).
Conidiophores carried on the aerial mycelium 25–65 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily phialides, often reduced to single phialides or as phialidic pegs; aerial phialides mono- and polyphialidic, subulate to subcylindrical, smooth- and thin-walled, 3–14 × 3–4 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: 6–10(–13) × 2–3 μm (av. 8 × 3 μm); 1-septate conidia: (9–)11–15(–17) × 2–4(–5) μm (av. 13 × 3 μm). Sporodochia pale rosy vinaceous to orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 3–9 × 2–3 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 8–12 × 2–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (1–)3–5-septate, hyaline, smooth- and thin-walled; 1-septate conidia: (14–)15–25(–32) × 2–4 μm (av. 20 × 3 μm); 2-septate conidia: (17–)19–25 × 3–4 μm (av. 22 × 4 μm); 3-septate conidia: (22–)30–40(–46) × (2–)3–4 μm (av. 35 × 4 μm); 4-septate conidia: (34–)36–40(–43) × 3–5 μm (av. 38 × 4 μm); 5-septate conidia: (36–)37–43(–50) × 3–5 μm (av. 40 × 4 μm). Chlamydospores not observed.
Culture characteristics — Colonies on PDA with an average radial growth rate of 2.6–3.4 mm/d at 24 °C. Colony surface pale rosy vinaceous grey, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse pale rosy vinaceous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, lacking chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant pale rosy vinaceous to orange sporodochia forming on the carnation leaves.
Additional materials examined. Zaire, from Elaeis sp., 1949, T. Gogoi, CBS 218.49 = NRRL 36359. – Unknown locality, from Elaeis guineensis, 1952, J. Fraselle, CBS 255.52 = NRRL 36386.
Notes — Fusarium elaeidis formed a highly-supported subclade in Clade III, closely related to F. callistephi, F. cugenangense and the untreated Fusarium clade. See notes under F. callistephi for distinguishing morphological features.
Fusarium fabacearum L. Lombard, Crous & Lampr., sp. nov. — MycoBank MB826839; Fig. 8
Fig. 8.
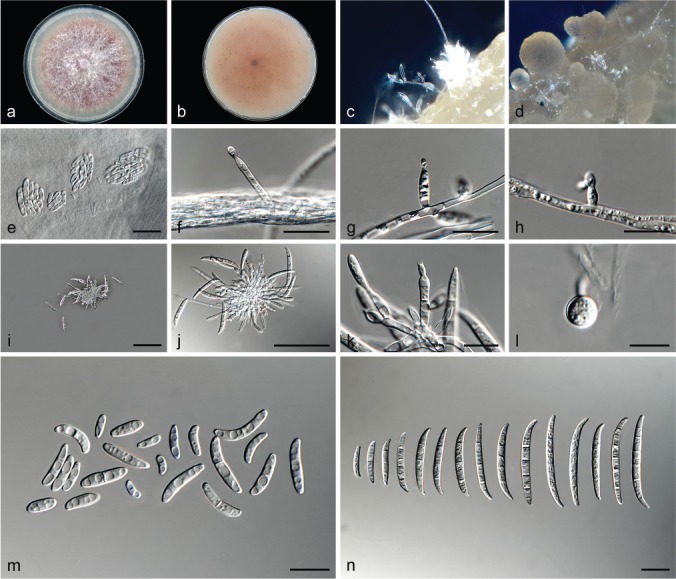
Fusarium fabacearum (ex-type culture CBS 144743). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c. conidiophores on surface of carnation leaf; d. sporodochia on carnation leaves; e. false head carried on a phialide on aerial mycelium; f–h. conidiophores and phialides on aerial mycelium; i–k. sporodochia and sporodochial conidiophores; l. chlamydospore; m. aerial conidia (microconidia); n. sporodochial conidia (macroconidia). — Scale bars: e–h, k–n = 10 μm; i–j = 50 μm.
Etymology. Name refers to the plant family, Fabaceae, which includes the plant host Glycine max from which this fungus was first isolated.
Typus. South Africa, North West Province, from Glycine max, 2010, S.C. Lamprecht (holotype CBS H-23613 designated here, culture ex-type CBS 144743 = CPC 25802).
Conidiophores carried on the aerial mycelium 25–50 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 11–15 × 3–4 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (4–)5–9(–13) × 2–3 μm (av. 7 × 3 μm); 1-septate conidia: (12–)13–15(–16) × 3–4 μm (av. 14 × 3 μm). Sporodochia pale luteous to orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 4–7 × 3 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 7–10 × 2–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (1–)3–4(–5)-septate, hyaline, smooth- and thin-walled; 1-septate conidia: (15–)16–24(–25) × 3–4 μm (av. 20 × 3 μm); 3-septate conidia: (24–)27–33(–36) × (2–)3–5 μm (av. 30 × 4 μm); 4-septate conidia: (32–)33–37(–40) × 3–5 μm (av. 35 × 4 μm); 5-septate conidia: (35–)38–44 × 3–4 μm (av. 41 × 4 μm). Chlamydospores globose to subglobose, formed terminally, 5–8 μm diam.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.0–4.4 mm/d at 24 °C. Colony surface pale vinaceous grey to vinaceous grey, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse pale vinaceous grey, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, with abundant chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant pale luteous to orange sporodochia forming on the carnation leaves.
Additional materials examined. South Africa, North West Province, from Glycine max, 2010, S.C. Lamprecht, CBS 144744 = CPC 25803; from Zea mays, 2008, C.M. Bezuidenhout, CBS 144742 = CPC 25801.
Notes — Fusarium fabacearum formed a highly-supported subclade in Clade III, closely related to F. carminascens and F. glycines. See notes under F. carminascens for distinguishing morphological features.
Fusarium glycines L. Lombard, Crous & Lampr., sp. nov. — MycoBank MB826840; Fig. 9
Fig. 9.
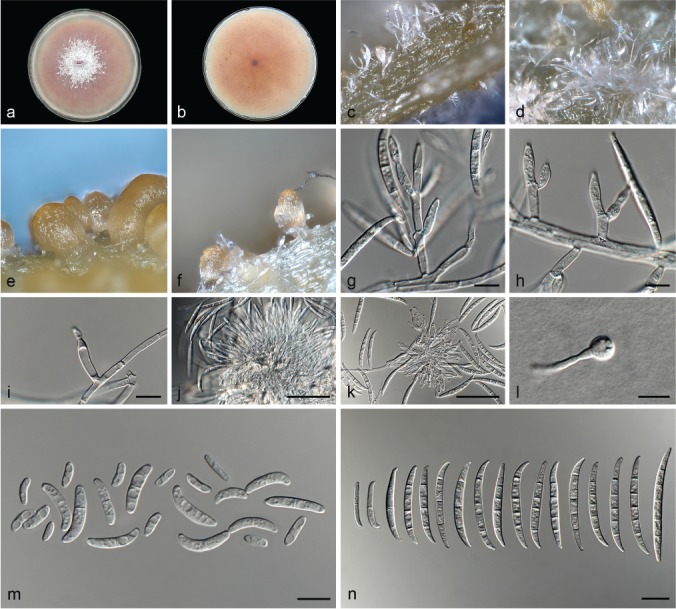
Fusarium glycines (ex-type culture CBS 144746). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. sporodochia on carnation leaves; g–i. conidiophores and phialides on aerial mycelium; j–k. sporodochia and sporodochial conidiophores; l. chlamydospore; m. aerial conidia (microconidia); n. sporodochial conidia (macroconidia). — Scale bars: g–i, l–n = 10 μm; j–k = 50 μm.
Etymology. Name refers to the plant genus Glycine from which this fungus was isolated.
Typus. South Africa, North West Province, from Glycine max, 2010, S.C. Lamprecht (holotype CBS H-23614 designated here, culture ex-type CBS 144746 = CPC 25808).
Conidiophores carried on the aerial mycelium 5–45 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 15–25 × 2–4 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: 7–11(–13) × 3–4 μm (av. 9 × 3 μm); 1-septate conidia: (13–)14–16(–18) × 3–4 μm (av. 15 × 3 μm). Sporodochia bright orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thinwalled stipe, 4–9 × 2–4 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 12–14 × 2–5 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (1–)3–5-septate, hyaline, smooth- and thin-walled; 1-septate conidia: 20–25 × 3–4 μm (av. 23 × 3 μm); 3-septate conidia: 37–43(–48) × 4–5 μm (av. 38 × 4 μm); 4-septate conidia: 44–46(–51) × 4–5 μm (av. 42 × 4 μm); 5-septate conidia: 43–49(–52) × 4–5 μm (av. 46 × 4 μm).
Chlamydospores globose to subglobose, formed terminally, 4–8 μm diam.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.0–4.4 mm/d at 24 °C. Colony surface vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse vinaceous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, with abundant chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant bright orange sporodochia forming on the carnation leaves.
Additional materials examined. Argentina, substrate unknown, date unknown, C.J.M. Carrera, CBS 214.49 = NRRL 36356 = LCF F-245. – Italy, from Ocimum basilicum, 1989, G. Tamiette & A. Matta, CBS 200.89. – South Africa, North West Province, from Glycine max, 2010, S.C. Lamprecht, CBS 144745 = CPC 25804. – Unknown locality, from Linum usitatissium, 1933, E.C. Stakman, CBS 176.33 = NRRL 36286.
Notes — Fusarium glycines formed a highly-supported subclade in Clade III, closely related to F. carminascens and F. fabacearum. See notes under F. carminascens for distinguishing morphological features.
Fusarium gossypinum L. Lombard & Crous, sp. nov. — MycoBank MB826841; Fig. 10
Fig. 10.
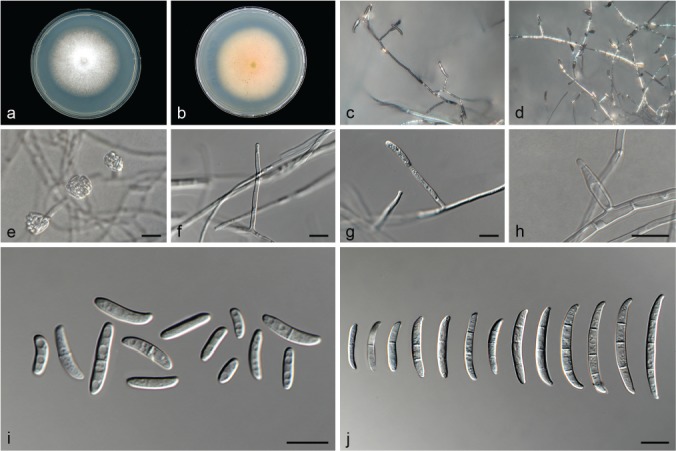
Fusarium gossypinum (ex-type culture CBS 116613). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e. false head carried on a phialide on aerial mycelium; f–h. conidiophores and phialides on aerial mycelium; i. aerial conidia (microconidia); j. sporodochial conidia (macroconidia). — Scale bars: e = 20 μm; f–j = 10 μm.
Etymology. Name refers to the plant genus Gossypium from which this fungus was isolated.
Typus. Ivory Coast, Bouaké, wilted Gossypium hirsutum, Sept. 1995, K. Abo (holotype CBS H-23615 designated here, culture ex-type CBS 116613).
Conidiophores carried on the aerial mycelium 35–75 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 3–30 × 2–4 μm, periclinal thickening inconspicuous or absent. Microconidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (5–)6–8(–11) × 2–4 μm (av. 7 × 3 μm); 1-septate conidia: (11–)12–14(–15) × 2–4 μm (av. 15 × 3 μm). Macroconidia also formed by phialides on aerial mycelium, falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (1–)3-septate, hyaline, smooth- and thin-walled; 1-septate conidia: 16–18 × 3 μm (av. 17 × 3 μm); 2-septate conidia: 21–23 × 3–4 μm (av. 22 × 3 μm); 3-septate conidia: (24–)27–35(–38) × 3–4 μm (av. 31 × 4 μm). Sporodochia absent. Chlamydospores not observed.
Culture characteristics — Colonies on PDA with an average radial growth rate of 1.6–2.8 mm/d at 24 °C. Colony surface white to pale rosy vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse pale rosy vinaceous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, lacking chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse lacking sporodochia on the carnation leaves.
Additional materials examined. Ivory Coast, Bouaké, wilted Gossypium hirsutum, Sept. 1995, K. Abo, CBS 116611 and CBS 116612.
Notes — Fusarium gossypinum formed a unique highly-supported subclade in Clade III. This species failed to produce any sporodochia on the carnation leaf pieces, but still produced abundant 3-septate macroconidia on the aerial mycelium. Other species included in Clade III, all readily produced sporodochia on carnation leaves.
Fusarium hoodiae L. Lombard, Crous & Lampr., sp. nov. — MycoBank MB826842; Fig. 11
Fig. 11.
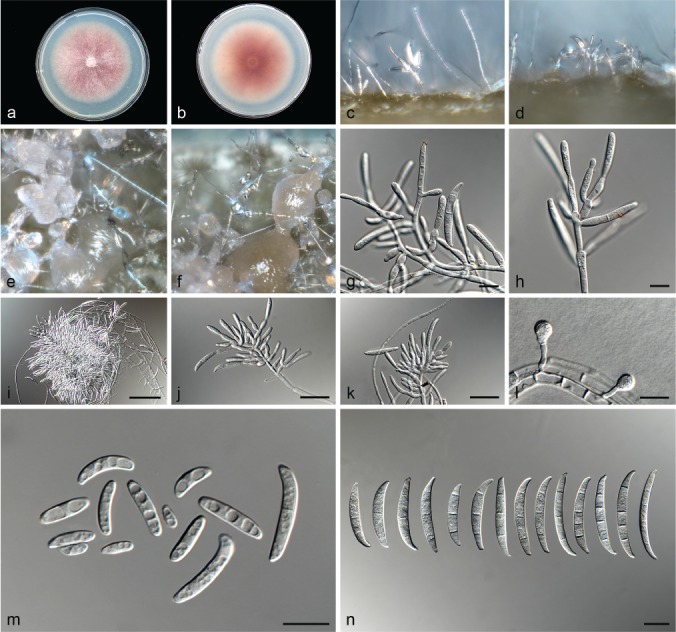
Fusarium hoodiae (ex-type culture CBS 132474). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. sporodochia on carnation leaves; g–h. conidiophores and phialides on aerial mycelium; i–k. sporodochia and sporodochial conidiophores; l. chlamydospore; m. aerial conidia (microconidia); n. sporodochial conidia (macroconidia). — Scale bars: g–h, l–n = 10 μm; i = 50 μm; j–k = 20 μm.
Etymology. Name refers to the plant genus Hoodia from which this fungus was isolated.
Typus. South Africa, Northern Cape Province, Prieska, root of Hoodia gordonii, 2002, O.A. Philippou (holotype CBS H-23616 designated here, culture ex-type CBS 132474).
Conidiophores carried on the aerial mycelium 40–60 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 15–24 × 2–3 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (5–)6–10(–16) × 2–4 μm (av. 8 × 3 μm); 1-septate conidia: (11–)12–16(–17) × 3–4 μm (av. 14 × 3 μm). Sporodochia pale vinaceous to light orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 7–11 × 3–5 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 7–13 × 2–5 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (1–)3(–4)-septate, hyaline, smooth- and thin-walled; 1-septate conidia: 20–33 × 3–5 μm (av. 25 × 4 μm); 3-septate conidia: (20–)27–39(–45) × 3–5 μm (av. 33 × 4 μm); 4-septate conidia: (35–)36–46(–51) × 4–5 μm (av. 41 × 5 μm). Chlamydospores globose to subglobose, formed terminally, 4–11 μm diam.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.1–4.5 mm/d at 24 °C. Colony surface pale vinaceous grey to livid vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse livid purple to pale vinaceous grey, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, with abundant chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant pale vinaceous to light orange sporodochia forming on the carnation leaves.
Additional materials examined. South Africa, Northern Cape Province, Prieska, root of Hoodia gordonii, 2002, O.A. Philippou, CBS 132476, CBS 132477.
Notes — Fusarium hoodiae formed a weakly supported clade constituting Clade IV in this phylogenetic study. All three isolates studied here, produced pale vinaceous to pale orange sporodochia on the carnation leaf pieces, unique for all the isolates studied.
Fusarium languescens L. Lombard & Crous, sp. nov. — MycoBank MB826843; Fig. 12
Fig. 12.
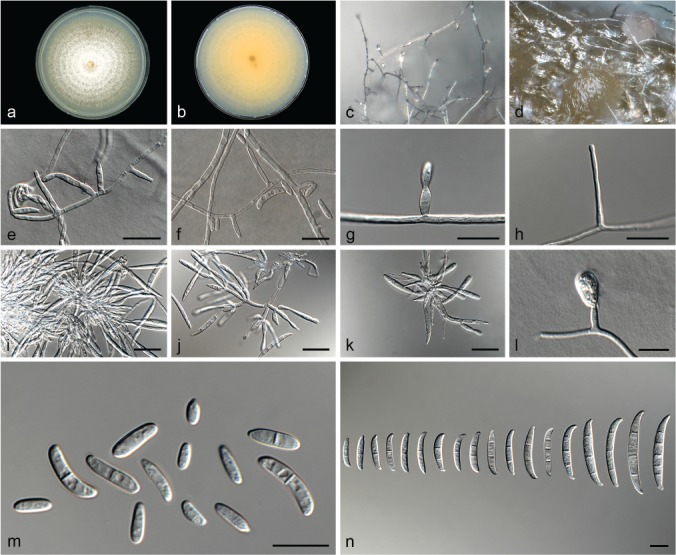
Fusarium languescens (ex-type culture CBS 645.78). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c. conidiophores on surface of carnation leaf; d. sporodochia on carnation leaves; e–h. conidiophores and phialides on aerial mycelium; i–k. sporodochia and sporodochial conidiophores; l. chlamydospore; m. aerial conidia (microconidia); n. sporodochial conidia (macroconidia). — Scale bars: e–h, l–n = 10 μm; i–k = 20 μm.
Etymology. Name refers to the wilting symptoms associated with infections of this fungus.
Typus. Morocco, Solanum lycopersicum, date and collector unknown (holo-type CBS H-23617 designated here, culture ex-type CBS 645.78 = NRRL 36531).
Conidiophores carried on the aerial mycelium 25–30 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 7–22 × 2–4 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (4–)5–9(–12) × 2–3 μm (av. 7 × 3 μm); 1-septate conidia: (9–)11–15 × 2–4 μm (av. 13 × 3 μm). Sporodochia light orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 5–10 × 3–4 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 10–14 × 2–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, 1–3(–5)-septate, hyaline, smooth- and thin-walled; 1-septate conidia: (15–)18–23(–30) × 3–4 μm (av. 20 × 3 μm); 2-septate conidia: (14–)16–22(–24) × 4 μm (av. 19 × 3 μm); 3-septate conidia: (22–)26–38(–47) × 3–5 μm (av. 32 × 4 μm); 5-septate conidia: 32–40 × 4–5 μm (av. 36 × 5 μm). Chlamydospores globose to subglobose, formed terminally, 6–9 μm diam.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.1–4.5 mm/d at 24 °C. Colony surface flesh to rosy vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse pale luteous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, with abundant chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant light orange sporodochia forming on the carnation leaves.
Additional materials examined. Israel, Bet Dagan, Solanum lycopersicum, 1986, R. Cohn, CBS 413.90 = ATCC 66046 = NRRL 36465. – Morocco, Solanum lycopersicum, date and collector unknown, CBS 646.78 = NRRL 36532. – Netherlands, Solanum lycopersicum, 1991, D.H. Elgersma, CBS 300.91 = NRRL 36416, CBS 302.91 = NRRL 36419. – South Africa, Zea mays, date and collector unknown, CBS 119796 = MRC 8437. – Unknown locality, Solanum lycopersicum, date and collector unknown, CBS 872.95 = NRRL 36570.
Notes — Fusarium languescens forms the highly-supported Clade VI, which mostly includes strains associated with tomato wilt. This species displays morphological overlap with several species treated here. Therefore, phylogenetic inference is needed to accurately identify this species.
Fusarium libertatis L. Lombard, Crous, sp. nov. — MycoBank MB826844; Fig. 13
Fig. 13.
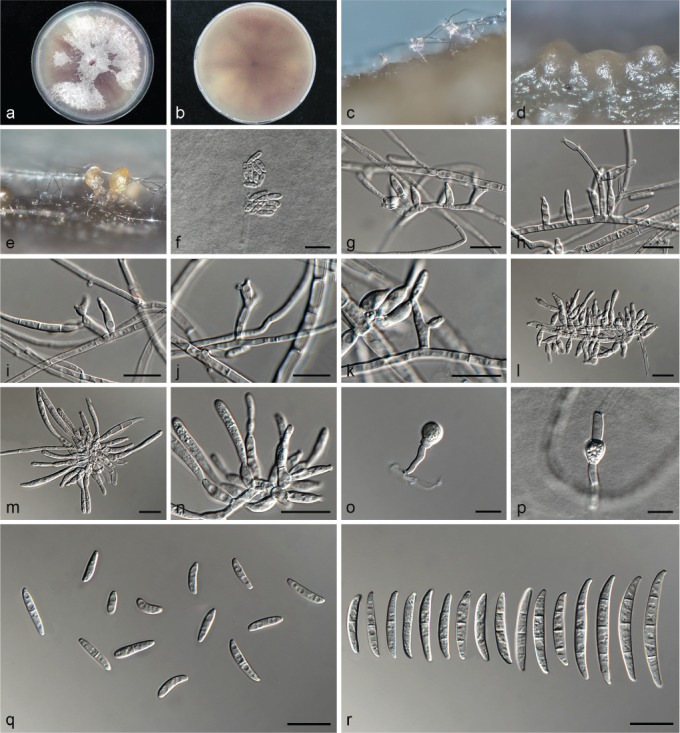
Fusarium libertatis (ex-type culture CBS 144749). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–e. conidiophores on surface of carnation leaf; g–k. conidiophores and phialides on aerial mycelium; g–h. monophialides; i–k. polyphialides; l–n. sporodochia and sporodochial conidiophores; n. phialides of sporodochial conidiophores; o–p. chlamydospores; q. aerial conidia (microconidia); r. sporodochial conidia (macroconidia). — Scale bars: c–r = 10 μm.
Etymology. Name refers to ‘freedom’. Fusarium libertatis was isolated from the rock surfaces in the stone quarry on Robben Island where the prisoners were forced to work. It is named in remembrance of all those who through the centuries were incarcerated on the Island for their different political beliefs.
Typus. South Africa, Western Cape Province, Robben Island, Van Riebeeck’s Quarry, from rock surfaces, May 2015, P.W. Crous (holotype CBS H-23618 designated here, culture ex-type CBS 144749 = CPC 28465).
Conidiophores carried on the aerial mycelium 2–30 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily phialides, often reduced to single phialides; aerial phialides mono- and polyphialidic, subulate to subcylindrical, smooth- and thin-walled, 8–13 × 2–4 μm, sometimes proliferating percurrently, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (6–)7–9(–11) × 2–4 μm (av. 8 × 3 μm); 1-septate conidia: (11–)12–14(–15) × 2–4 μm (av. 13 × 3 μm). Sporodochia bright orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 4–8 × 3–4 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 6–12 × 2–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, 1–3-septate, hyaline, smooth- and thin-walled; 1-septate conidia: (15–)17–21(–23) × 2–4 μm (av. 19 × 3 μm); 2-septate conidia: (18–)20–24(–25) × 2–3(–4) μm (av. 22 × 4 μm); 3-septate conidia: (24–)30–38(–40) × (2–)3–5 μm (av. 34 × 4 μm). Chlamydospores globose to subglobose, formed terminally and intercalarily, carried singly, 5–9 μm diam.
Culture characteristics — Colonies on PDA with an average radial growth rate of 2.3–4.4 mm/d at 24 °C. Colony surface vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse vinaceous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, with abundant chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant bright orange sporodochia forming on the carnation leaves.
Additional materials examined. South Africa, Western Cape Province, from Aspalathus sp., 2008, C.M. Bezuidenhout, CBS 144747 = CPC 25788, CBS 144748 = CPC 25782.
Notes — Fusarium libertatis formed a unique well-supported clade Clade (II). This species readily produced polyphialidic conidiogenous cells on its aerial mycelium and can be distinguished from the other species (F. carminascens, F. curvatum and F. elaeidis) found to produce polyphialides by only producing up to 3-septate macroconidia, whereas the other polyphialidic species produce up to 5-septate macroconidia.
Fusarium nirenbergiae L. Lombard & Crous, sp. nov. — MycoBank MB826845; Fig. 14
Fig. 14.
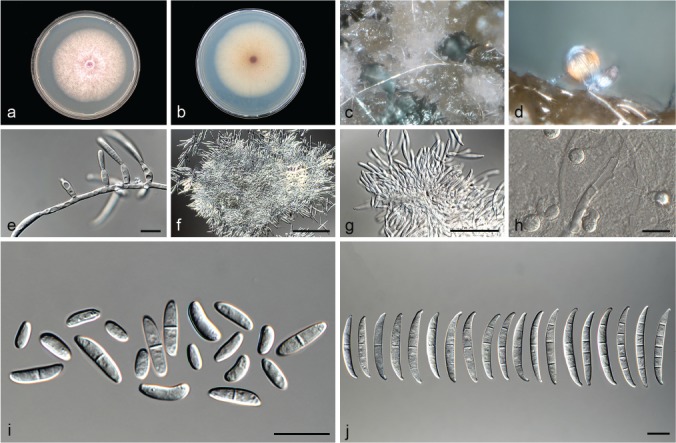
Fusarium nirenbergiae (ex-type culture CBS 840.88). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c. conidiophores on surface of carnation leaf; d. sporodochia on carnation leaves; e. conidiophores and phialides on aerial mycelium; f–g. sporodochia and sporodochial conidiophores; h. chlamydospore; i. aerial conidia (microconidia); j. sporodochial conidia (macroconidia). — Scale bars: e, h–j = 10 μm; f–g = 50 μm.
Etymology. Named in honour of Prof. H.I. Nirenberg for her contribution to our understanding of Fusarium taxonomy.
Typus. Netherlands, Aalsmeer, from Dianthus caryophyllus, 1988, H. Rattink (holotype CBS H-23619 designated here, culture ex-type CBS 840.88).
Conidiophores carried on the aerial mycelium 18–50 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 8–24 × 2–4 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (5–)6–10(–11) × 2–4 μm (av. 8 × 3 μm); 1-septate conidia: (9–)10–14(–15) × 2–4 μm (av. 12 × 3 μm). Sporodochia bright orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 6–14 × 3–5 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 8–18 × 2–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, 1–5-septate, hyaline, smooth- and thin-walled; 1-septate conidia: 15–29(–34) × 3–4 μm (av. 22 × 4 μm); 2-septate conidia: (18–)19–31(–39) × 2–4(–5) μm (av. 25 × 3 μm); 3-septate conidia: (30–)32–40(–43) × 3–4 μm (av. 36 × 4 μm); 4-septate conidia: (34–)36–44(–48) × 3–5 μm (av. 40 × 4 μm); 5-septate conidia: (36–)43–59(–66) × 3–5 μm (av. 51 × 4 μm). Chlamydospores globose to subglobose, formed terminally, 4–6 μm diam.
Culture characteristics — Colonies on PDA with an average radial growth rate of 2.9–4.2 mm/d at 24 °C. Colony surface pale vinaceous to vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse pale vinaceous grey to greyish lilac, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, with abundant chlamydospores, aerial mycelium sparse with moderate sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant bright orange sporodochia forming on the carnation leaves.
Additional materials examined. Brazil, from Passiflora edulis, 1968, W. Gerlach, CBS 744.79 = BBA 62355 = NRRL 22549. – Italy, Napoli, Castellammare di Stabia, from Bouvardia longiflora, July 1986, B. Aloj, CBS 196.87 = NRRL 26219. – Netherlands, Berkel, from Solanum lycopersicum, 16 May 1968, G. Weststeijn, CBS 758.68 = NRRL 36546. – South Africa, Western Cape Province, Riebeeck-Wes, from Agathosma betulina, 2001, K. Lubbe, CBS 115424 = CPC 5312; Stellenbosch, Elsenberg farm, from Agathosma betulina, 2001, K. Lubbe, CBS 115416 = CPC 5307, CBS 115417 = CPC 5306, CBS 115419 = CPC 5308. – USA, California, from amputated human toe, unknown date and collector, CBS 130300 = NRRL 26368; Florida, from Solanum tuberosum, 1923, H.W. Wollenweber, CBS 181.32 = NRRL 36303; from Chrysanthemum sp., date unknown, G.M. Armstrong & J.K. Armstrong, CBS 127.81 = BBA 63924 = NRRL 36229; Florida, from Chrysanthemum sp., date unknown, A.W. Engelhard, CBS 129.81 = BBA 63926 = NRRL 22539; Maryland, Beltsville, from tulip roots, 1991, R.L. Lumsden, CBS 123062 = GJS 91-17; Florida, Immokalee, from Solanum lycopersicum, date unknown, J. Swezey, CBS 130303; Texas, San Antonio, from human leg ulcer, date and collector unknown, CBS 130301 = NRRL 26374. – Unknown locality, from Secale cereale, date unknown, H.W. Wollenweber, CBS 129.24; from Musa sp., date unknown, E.W. Mason, CBS 149.25 = NRRL 36261.
Notes — Fusarium nirenbergiae formed a well-supported subclade in Clade VIII, closely related to F. curvatum. See notes under F. curvatum for distinguishing morphological features.
Fusarium oxysporum Schltdl., Fl. Berol. 2: 139. 1824 — Fig. 15
Fig. 15.
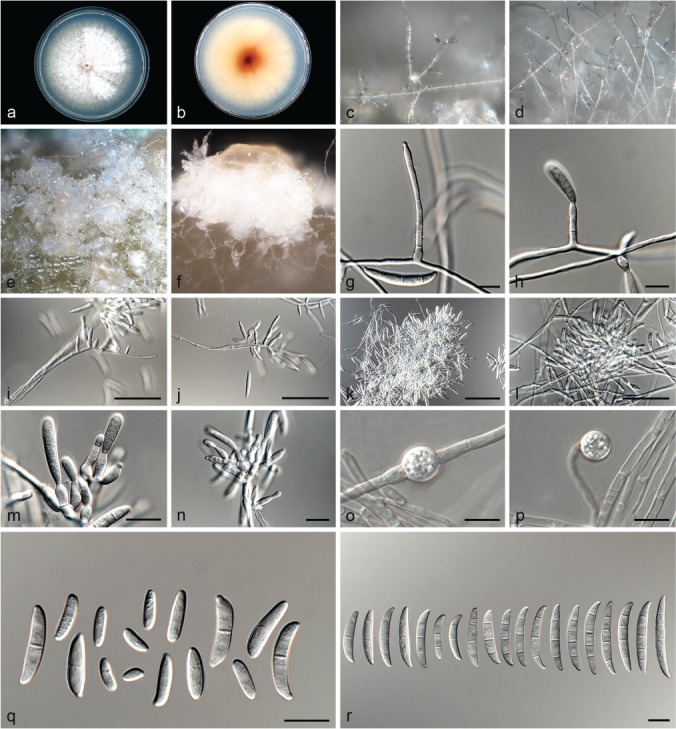
Fusarium oxysporum (ex-epitype culture CBS 144134). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. sporodochia on carnation leaves; g–j. conidiophores and phialides on aerial mycelium; k–n. sporodochia and sporodochial conidiophores; o–p. chlamydospores; q. aerial conidia (microconidia); r. sporodochial conidia (macroconidia). — Scale bars: g–h, m–r = 10 μm; i–l = 50 μm.
Synonyms. Fusarium bulbigenum Cooke & Massee, Grevillea 16: 49. 1887.
Fusarium vasinfectum G.F.Atk., Bull. Alabama Agric. Exper. Station 41: 19. 1892.
Fusarium dianthi Prill. & Delacr., Compt. Rend. Acad. Sci. 129: 744. 1899.
Fusarium lini Bolley, Proc. Ann. Meeting Soc. Prom. Agr. Sci. 21: 1–4. 1902.
Fusarium orthoceras Appel & Wollenw., Arb. Kaiserl. Biol. Anst. Ld.- u. Forstw. 8: 152. 1910.
Fusarium citrinum Wollenw., Maine Agric. Exp. Sta. Bull. 219: 256. 1913.
Fusarium angustum Sherb., Cornell Univ. Agric. Exp. Sta. Mem. 6: 203. 1915.
Fusarium lutulatum Sherb., Cornell Univ. Agric. Exp. Sta. Mem. 6: 209. 1915.
Fusarium bostrycoides Wollenw. & Reinking, Phytopathology 15: 166. 1925.
Diplosporium vaginae Nann., Atti Reale Accad. Fisiocrit. Siena sér. 4, 17: 491. 1926.
For additional synonyms see Index Fungorum and MycoBank.
Typification. Germany, Berlin, from rotten tuber of Solanum tuberosum, 1824, D.L.F. von Schlechtendal, HAL 1612 F, holotype in HAL; (epitype designated here: Germany, Berlin, from rotten tuber of Solanum tuberosum, 17 Oct. 2017, L. Lombard, epitype CBS H-23620, MBT382397, culture ex-epitype CBS 144134).
Conidiophores carried on the aerial mycelium 15–75 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 11–40 × 2–4 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (4–)6–10(–11) × 2–4 μm (av. 8 × 3 μm); 1-septate conidia: 13–15(–16) × 2–4 μm (av. 14 × 3 μm). Sporodochia bright orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 4–10 × 4–5 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 8–13 × 3–5 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (1–)3(–5)-septate, hyaline, smooth- and thin-walled; 1-septate conidia: (21–)22–26 × 4–5 μm (av. 24 × 4 μm); 2-septate conidia: 20–26(–27) × 4–5 μm (av. 23 × 4 μm); 3-septate conidia: (22–)25–29(–31) × 4–5 μm (av. 27 × 4 μm); 4-septate conidia: (30–)31–35 × 4–5 μm (av. 33 × 5 μm); 5-septate conidia: 35–38 × 5–6 μm (av. 37 × 5 μm). Chlamydospores globose to subglobose, formed intercalarily or terminally, 5–10 μm diam.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.0–4.0 mm/d at 24 °C. Colony surface pale vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse vinaceous to rosy vinaceous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, producing abundant chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant bright orange sporodochia forming on the carnation leaves.
Additional materials examined. Germany, from rotten tuber of Solanum tuberosum, 17 Oct. 2017, L. Lombard, CBS 144135. – South Africa, Western Cape Province, from Protea sp., date unknown, C.M. Bezuidenhout, CPC 25822. – South East Asia, from Camellia sinensis, 1949, F. Bugnicourt, CBS 221.49 = IHEM 4508 = NRRL 22546.
Notes — Fusarium oxysporum formed a well-supported subclade in Clade V with F. triseptatum as closest relative. Both species in Clade V displayed some morphological overlap. However, the 1-septate ((21–)22–26 × 4–5 μm (av. 24 × 4 μm) and 2-septate (20–26(–27) × 4–5 μm (av. 23 × 4 μm) macroconidia of F. oxysporum are larger than those of F. triseptatum ((18–)19–23(–24) × 3–4 μm (av. 20 × 3 μm) and 17–25(–26) × 3 μm (av. 21 × 3 μm), respectively), whereas the 3-septate ((25–)27–39(–47) × 4–5 μm (av. 33 × 3 μm)), 4-septate ((31–)34–40(–41) × 4–5 μm (av. 37 × 4 μm)) and 5-septate ((33–48(–49) × 4–5 μm (av. 40 × 4 μm)) macroconidia of F. triseptatum are larger than those of F. oxysporum ((22–)25–29(–31) × 4–5 μm (av. 27 × 4 μm), (30–)31–35 × 4–5 μm (av. 33 × 5 μm) and 35–38 × 5–6 μm (av. 37 × 5 μm), respectively). Additionally, all isolates of F. oxysporum produced abundant bright orange sporodochia on carnation leaf pieces, not observed for any of the F. triseptatum isolates studied.
Fusarium pharetrum L. Lombard & Crous, sp. nov. — MycoBank MB826846; Fig. 16
Fig. 16.
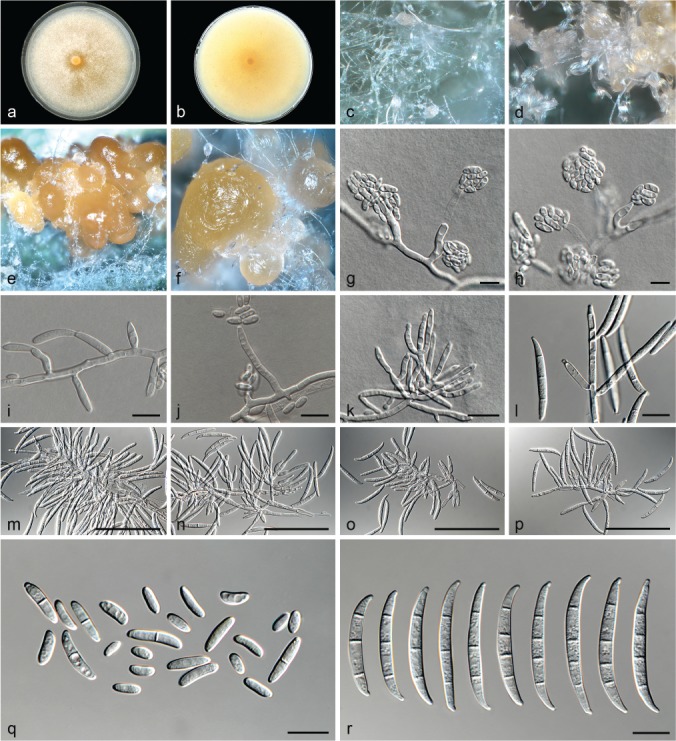
Fusarium pharetum (ex-type culture CBS 144751). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. sporodochia on carnation leaves; g–h. false heads carried on a phialide on aerial mycelium; i–l. conidiophores and phialides on aerial mycelium; m–p. sporodochia and sporodochial conidiophores; q. aerial conidia (microconidia); r. sporodochial conidia (macroconidia). — Scale bars: g–l, q–r = 10 μm; m–p = 50 μm.
Etymology. Name refers to the practice of the Southern African indigenous San people of hollowing out the tubular branches of the host plant, Aloidendron dichotomum, to form quivers (Latin pharetra) for their arrows.
Typus. South Africa, from Aliodendron dichotomum, 2000, F. van der Walt & G.J. Marais (holotype CBS H-23621 designated here, culture ex-type CBS 144751 = CPC 30824).
Conidiophores carried on the aerial mycelium 20–75 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 4–28 × 2–5 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: 5–9(–13) × 2–3 μm (av. 7 × 3 μm); 1-septate conidia: (10–)12–16(–18) × 2–4 μm (av. 14 × 3 μm). Sporodochia rosy vinaceous to orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 5–10 × 3–5 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 7–13 × 3–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, 3(–4)-septate, hyaline, smooth- and thin-walled; 3-septate conidia: (22–)27–35(–39) × 3–5 μm (av. 31 × 4 μm); 4-septate conidia: (34–)36–40(–41) × 3–5 μm (av. 36 × 5 μm). Chlamydospores not observed.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.1–4.5 mm/d at 24 °C. Colony surface rosy vinaceous, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse rosy vinaceous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, lacking chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant rosy vinaceous to orange sporodochia forming on the carnation leaves.
Additional material examined. South Africa, from Aliodendron dichotomum, 2000, F. van der Walt & G.J. Marais, CBS 144750 = CPC 30822.
Notes — Fusarium pharetrum formed a well-supported subclade in Clade VII, closely related to F. contaminatum and F. veterinarium. See notes under F. contaminatum for distinguishing morphological features.
Fusarium triseptatum L. Lombard & Crous, sp. nov. — MycoBank MB826847; Fig. 17
Fig. 17.
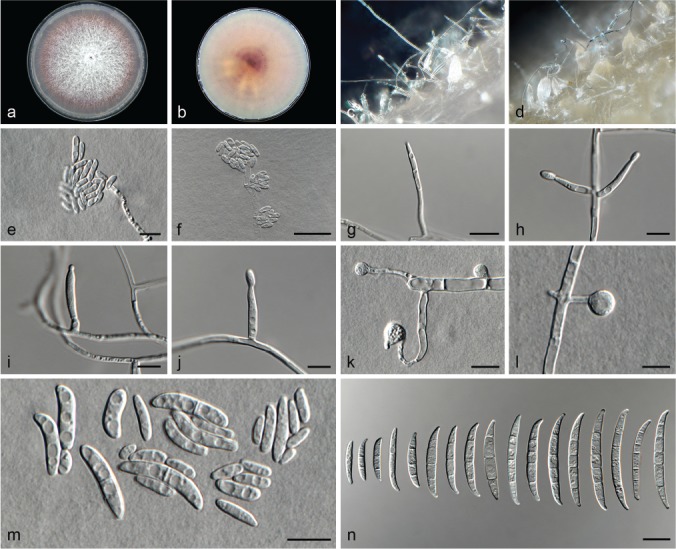
Fusarium triseptatum (ex-type culture CBS 258.50). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. false heads carried on a phialide on aerial mycelium; g–j. conidiophores and phialides on aerial mycelium; k–l. chlamydospores; m. microconidia; n. macroconidia. — Scale bars: e, g–n = 10 μm; f = 20 μm.
Etymology. Name refers to the abundant 3-septate macroconidia produced by this fungus.
Typus. USA, locality unknown, from Ipomoea batatas, 1950, T.T. McClure (holotype CBS H-23622 designated here, culture ex-type CBS 258.50 = NRRL 36389).
Conidiophores carried on the aerial mycelium 5–40 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 6–22 × 2–4 μm, periclinal thickening inconspicuous or absent. Microconidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (5–)6–10(–13) × 1–3 μm (av. 8 × 3 μm); 1-septate conidia: (12–)14–16(–18) × 2–4 μm (av. 15 × 3 μm). Macroconidia also formed by phialides on aerial mycelium, falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, (1–)3(–5)-septate, hyaline, smooth- and thin-walled; 1-septate conidia: (18–)19–23(–24) × 3–4 μm (av. 20 × 3 μm); 2-septate conidia: 17–25(–26) × 3 μm (av. 21 × 3 μm); 3-septate conidia: (25–)27–39(–47) × 4–5 μm (av. 33 × 3 μm); 4-septate conidia: (31–)34–40(–41) × 4–5 μm (av. 37 × 4 μm); 5-septate conidia: 33–48(–49) × 4–5 μm (av. 40 × 4 μm). Sporodochia absent. Chlamydospores globose to subglobose, formed terminally, 5–12 μm diam.
Culture characteristics — Colonies on PDA with an average radial growth rate of 2.2–3.4 mm/d at 24 °C. Colony surface pale vinaceous grey to vinaceous grey, floccose with abundant aerial mycelium; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse pale vinaceous grey, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, with abundant chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse lacking sporodochia on the carnation leaves.
Additional materials examined. Ivory Coast, Béoumi, wilted Gossypium hirsutum, Oct. 1996, K. Abo, CBS 116619. – Papua New Guinea, Suki village, from sago starch, 2005, A. Greenhill, CBS 119665. – USA, Tennessee, from human eye, collector and date unknown, CBS 130302 = NRRL 26360 = FRC 755.
Notes — Fusarium triseptatum formed a highly-supported subclade in Clade V, closely related to F. oxysporum. See notes under F. oxysporum for distinguishing morphological features.
Fusarium veterinarium L. Lombard & Crous, sp. nov. — MycoBank MB826849; Fig. 18
Fig. 18.
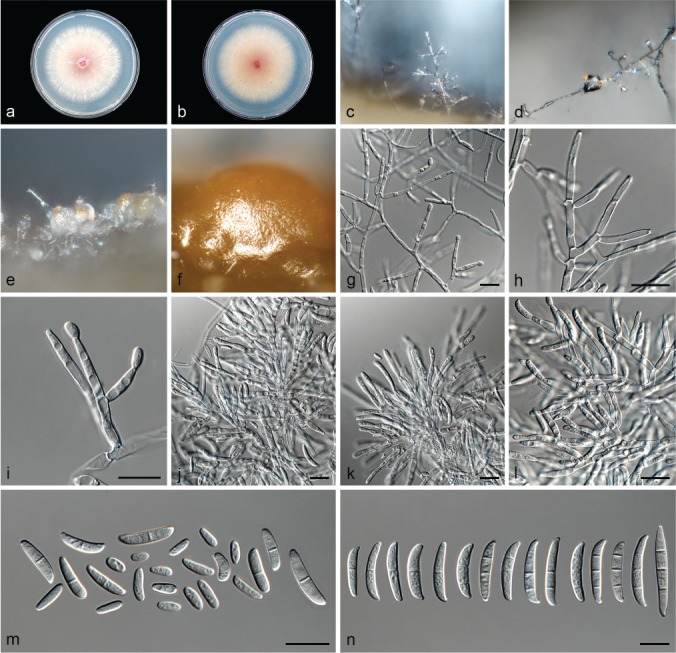
Fusarium veterinarium (ex-type culture CBS 109898). a–b. Colony on PDA; a. surface of colony on PDA after 7 d at 24 °C under continuous white light; b. reverse of colony on PDA; c–d. conidiophores on surface of carnation leaf; e–f. sporodochia on carnation leaves; g–i. conidiophores and phialides on aerial mycelium; j–l. sporodochia and sporodochial conidiophores; m. aerial conidia (microconidia); n. sporodochial conidia (macroconidia). — Scale bars: g–n = 10 μm.
Etymology. Name refers to the fact that this fungus was isolated mostly from veterinary samples.
Typus. Netherlands, from shark peritoneum, date unknown, C. Hoek (holotype CBS H-23623 designated here, culture ex-type CBS 109898 = NRRL 36153).
Conidiophores carried on the aerial mycelium 12–90 μm tall, unbranched or sparingly branched, bearing terminal or intercalarily monophialides, often reduced to single phialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, 8–24 × 2–4 μm, periclinal thickening inconspicuous or absent; aerial conidia forming small false heads on the tips of the phialides, hyaline, ellipsoidal to falcate, smooth- and thin-walled, 0–1-septate; 0-septate conidia: (4–)6–8(–11) × 2–4 μm (av. 7 × 3 μm); 1-septate conidia: (9–)10–14(–15) × 2–4 μm (av. 12 × 3 μm). Sporodochia bright orange, formed abundantly on carnation leaves. Conidiophores in sporodochia verticillately branched and densely packed, consisting of a short, smooth- and thin-walled stipe, 8–13 × 3–4 μm, bearing apical whorls of 2–3 monophialides or rarely as single lateral monophialides; sporodochial phialides subulate to subcylindrical, 10–15 × 2–4 μm, smooth- and thin-walled, sometimes showing a reduced and flared collarette. Sporodochial conidia falcate, curved dorsiventrally with almost parallel sides tapering slightly towards both ends, with a blunt to papillate, curved apical cell and a blunt to foot-like basal cell, 1–(2–)3-septate, hyaline, smooth- and thin-walled; 1-septate conidia: (12–)15–19(–20) × 3–4 μm (av. 17 × 3 μm); 2-septate conidia: (16–)17–21(–24) × 3–4 μm (av. 19 × 3 μm); 3-septate conidia: (19–)20–24(–27) × 3–4 μm (av. 22 × 3 μm). Chlamydospores not observed.
Culture characteristics — Colonies on PDA with an average radial growth rate of 3.1–4.5 mm/d at 24 °C. Colony surface pale vinaceous grey, floccose with moderate aerial mycelium appearing wet; colony margins irregular, lobate, serrate or filiform. Odour absent. Reverse pale vinaceous, lacking diffusible pigment. On SNA, hyphae hyaline, smooth-walled, lacking chlamydospores, aerial mycelium sparse with abundant sporulation on the medium surface. On CLA, aerial mycelium sparse with abundant orange sporodochia forming on the carnation leaves.
Additional materials examined. Netherlands, from swab sample near filling apparatus, Apr. 2005, J. Houbraken, CBS 117787, CBS 117790; from pasteurized milk-based product, Apr. 2005, J. Houbraken, CBS 117791, CBS 117792. – USA, California, from endoscope of veterinary clinic, date and collector unknown, NRRL 62545; from canine stomach, date and collector unknown, NRRL 62547; Massachusetts, from mouse mucosa, date and collector unknown, NRRL 54984; from little blue penguin foot, date and collector unknown, NRRL 54996; Texas, from unknown animal faeces, date and collector unknown, NRRL 62542.
Notes — Fusarium veterinarium formed a highly-supported subclade in Clade VII, closely related to F. contaminatum and F. pharetrum. See notes under F. contaminatum for distinguishing morphological features.
DISCUSSION
Fusarium taxonomy and the underlying phylogenetic backbone on which it is based, is undergoing continuous revision. In modern day fungal taxonomy, phylogenetic inference plays a vital role to resolve the identity of cryptic species due to the paucity of morphological features. However, a key component of a robust phylogeny is the availability of living ex-type material to serve as basic reference point or ‘phylogenetic anchor’ on which comparative taxonomy can be based (Booth 1975). Epi- and/or neotypification provides a vital means where upon stability can be enforced into a chaotic classification system as being applied to F. oxysporum today.
Snyder & Hansen’s (1940) treatment of the section Elegans to represent only F. oxysporum, has resulted in a much too broad definition of this species. Based on this, the current morphological characters used to define F. oxysporum include aseptate microconidia forming false heads on short monophialides, commonly 3-septate macroconidia formed on monophialides or branched conidiophores in sporodochia, and chlamydospores that are either formed abundantly and quickly or slowly with some strains not forming them at all (Leslie & Summerell 2006, Fourie et al. 2011). In this study, all isolates were found to produce not only aseptate microconidia, but abundant 1-septate microconidia, all of which were carried on false heads. Several species were also found to form polyphialides (e.g., F. carminascens, F. curvatum, F. elaeidis and F. libertatis), a characteristic not associated with F. oxysporum morphology (Gerlach & Nirenberg 1982, Nelson et al. 1983, Leslie & Summerell 2006). Additionally, the majority of the species introduced here produced 4- to 5-septate macroconidia in the same abundance as the 3-septate macroconidia. Gerlach & Nirenberg (1982) also indicated the presence of 7-septate macroconidia, but these were not observed in this study given the media and growth conditions we employed. The ex-epitype strain of F. oxysporum designated here, agrees well with the morphological characteristics described by Wollenweber & Reinking (1935), Booth (1971), Gerlach & Nirenberg (1982) and Nelson et al. (1983). This strain produced abundant aseptate and 1-septate microconidia on monophialides only, abundant 3-septate macroconidia with much fewer 1-, 2-, 4- and 5-septate macroconidia on its sporodochia, and smooth-walled globose chlamydospores carried intercalarily and/or terminally. Although this strain was isolated from a potato tuber displaying symptoms of dry rot, the ability of this strain to induce these symptoms requires further investigation. Comparisons of the 15 novel Fusarium taxa introduced here, revealed subtle morphological distinctions between the species.
Fusarium carminascens, F. curvatum, F. elaeidis and F. libertatis readily formed polyphialides on the aerial mycelium, a feature not known for F. oxysporum (Wollenweber & Reinking 1935, Booth 1971, Gerlach & Nirenberg 1982, Nelson et al. 1983, Leslie & Summerell 2006). These four species are further distinguished from each other by the degree of septation and curvature of their macroconidia. Both F. carminascens and F. libertatis readily formed chlamydospores in culture, whereas no chlamydospores were observed for F. curvatum and F. elaeidis. Furthermore, all strains of F. carminascens produced an almost carmine red exudate on the aerial mycelium on PDA, not observed for any other strains studied here. The strong curvature of the macroconidia of F. curvatum is also a unique feature.
The remaining 11 novel species introduced here can be distinguished based on the degree of septation and dimensions of the macroconidia and the formation of chlamydospores in culture. Of these, F. contaminatum, F. gossypinum, F. hoodiae, F. languescens, F. pharetrum, F. triseptatum and F. veterinarium displayed some morphological overlap with the ex-epitype strain of F. oxysporum. However, F. contaminatum, F. gossypinum, F. pharetrum and F. veterinarium did not form chlamydospores in culture. These four species are easily distinguished based on macroconidial dimensions with F. contaminatum and F. veterinarium producing the smallest macroconidia. Fusarium hoodiae, F. languescens and F. triseptatum readily formed chlamydospores in culture and can be distinguished from each other and F. oxysporum based on their sporodochia. All strains of F. triseptatum failed to produce any sporodochia on the carnation leaf pieces, whereas F. hoodiae formed distinct pale vinaceous to pale orange sporodochia compared to the only pale orange sporodochia of F. languescens. Fusarium callistephi, F. fabacearum, F. glycines and F. nirenbergiae are easily distinguished from each other and F. oxysporum by the degree of macroconidial septation and dimensions. However, these subtle morphological differences need to be supported by phylogenetic inference to accurately discriminate between these novel species introduced in the FOSC in this study.
Individual analyses of the partial sequences of the four gene regions (cmdA, rpb2, tef1 and tub2) included in this study (results not shown) revealed that the tef1 gene region provided the best resolution to discriminate the novel species introduced here. The rpb2 gene region also provided good resolution, but with lower statistical support, whereas the cmdA and tub2 provided little to no support. However, the addition of the latter two gene regions to either or both the rpb2 and tef1 greatly increased the statistical support of each Clade (I–VIII) and their underlining subclades. Genealogical concordance phylogenetic species recognition analyses also indicated that there was no evidence of recombination detected between any of the Clades and subclades resolved in this study. Analysis of the IGS gene region (results not shown) provided contradictory tree topologies and support values, with several strains in Clades III, VII and VIII forming single lineages. Although O’Donnell et al. (2015) advocates the use of rpb1, rpb2 and tef1 for sequence-based identification of Fusarium species, attempts to generate rpb1 sequence data in this study failed for the majority of strains included in this study.
Previous studies of FOSC revealed a high phylogenetic diversity within this complex, resolving three (O’Donnell et al. 1998, Brankovics et al. 2017), four (O’Donnell et al. 2004) and five (Laurence et al. 2012) phylogenetic clades, respectively. Comparisons of all these clades with those resolved in this study, revealed that Clade I in this study correlates well with Clade 1 resolved by O’Donnell et al. (1998, 2004), Laurence et al. (2012) and Brankovics et al. (2017). Similarly, Clade VIII in this study matched with Clade 3 of each of these studies. Clade III correlated with Clade 2 resolved by O’Donnell et al. (2004) and Brankovics et al. (2017), and Clade V correlated with clades 4 and 5 of Laurence et al. (2012), and Clade 4 of O’Donnell et al. (2004). Clades II, IV, VI and VII resolved in this study did not match any of the clades resolved in these previous studies.
Comparisons of the origin of the strains studied here revealed some correlation within most of the Clades (and subclades). All veterinarian strains included in this study clustered together with some strains originating from equipment used in food processing in a highly-supported subclade representing F. veterinarium. Similarly, three strains collected from contaminated dairy products and fruit juice clustered together in the highly-supported (sub)clade representing F. contaminatum. The majority of the isolates collected from tomato (Solanum lycopersicum) also cluster together in a clade representing F. languescens, with a few clustering in the F. nirenbergiae (sub)clade. In contrast to these few highlighted examples, all medically related strains clustered in various well- to highly supported clades, representing F. cugenangense, F. nirenbergiae, F. triseptatum and the untreated Fusarium clade. The highest host/substrate diversity was found in the F. nirenbergiae (sub)clade which included several special forms in addition to the medically related strains.
The application of the special form and pathotype classification system can only be successfully applied if the species boundaries are well established (Woudenberg et al. 2015), which is clearly not the case within the FOSC. For the FOSC, special forms are defined by the accessory chromosome obtained via horizontal gene transfer, and the pathotype on the type of virulence genes carried by this chromosome and should not be confused with the species boundaries within the FOSC. Therefore, epitypification of F. oxysporum in this study has resulted in the recognition of 21 phylogenetic species of which 15 are provided with names here. Although this study includes only a small subset of strains belonging to the FOSC, the inclusion of more isolates will provide a much better perspective on the cryptic diversity within this important species complex, allowing additional species to be recognised. Furthermore, it is hoped that with the epitypification of F. oxysporum, the confusing and sometimes complicated subspecific classification systems that have been applied to the FOSC in the past will become obsolete and be replaced by a more stable and convenient species-level classification system. We believe that such a system will allow for better communication between Fusarium researchers in the medical, environmental and phytopathological fields.
Acknowledgements
The authors thank the technical staff, A. van Iperen, D. Vos-Kleyn and Y. Vlug for their valuable assistance with cultures.
REFERENCES
- Abd-Elsalam K, Khalil M, Aly AH, et al. 2002. Genetic diversity among Fusarium oxysporum f. sp. vasinfectum isolates revealed by UP-PCR and AFLP markers. Phytopathologia Mediterrianae 41: 252–258. [Google Scholar]
- Abd-Elsalam KA, Asran-Amal A, Schnieder F, et al. 2006. Molecular detection of Fusarium oxysporum f. sp. vasinfectum in cotton roots by PCR and real-time PCR assay. Journal of Plant Disease and Protection 113: 14–19. [Google Scholar]
- Abd-Elsalam KA, Omar MR, Migheli Q, et al. 2004. Genetic characterization of Fusarium oxysporum f. sp. vasinfectum isolates by random amplification of polymorphic DNA (RAPD) and amplified fragment length polymorphism (AFLP). Journal of Plant Diseases and Protection 111: 534–544. [Google Scholar]
- Abo K, Klein KK, Edel-Hermann V, et al. 2005. High genetic diversity among strains of Fusarium oxysporum f. sp. vasinfectum from cotton in Ivory Coast. Phytopathology 95: 1391–1396. [DOI] [PubMed] [Google Scholar]
- Adame-García J, Rodríguez-Guerra R, Iglesias-Andreu LG, et al. 2015. Molecular identification and pathogenic variation of Fusarium species isolated from Vanilla planifolia in Papantla Mexico. Botanical Sciences 93: 669–678. [Google Scholar]
- Aguayo J, Mostert D, Fourrier-Jeandel C, et al. 2017. Development of a hydrolysis probe-based real-time assay for the detection of tropical strains of Fusarium oxysporum f. sp. cubense race 4. PLoS ONE 12 (2): e0171767. doi: 10.1371/journal.pone.0171767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn IP, Chung HS, Lee Y-H. 1998. Vegetative compatibility groups and pathogenicity among isolates of Fusarium oxysporum f. sp. cucumerinum. Plant Disease 82: 244–246. [DOI] [PubMed] [Google Scholar]
- Al-Hatmi AMS, Bonifaz A, Ranque S, et al. 2018. Current antifungal treatment of fusariosis. International Journal of Antimicrobial Agents 51: 326–332. [DOI] [PubMed] [Google Scholar]
- Al-Husien N, Hamwieh A, Ahmed S, et al. 2017. Genetic diversity of Fusarium oxysporum f. sp. lentis population affecting lentil in Syria. Journal of Phytopathology 165: 306–312. [Google Scholar]
- Alexander LJ, Tucker CM. 1945. Physiological specialization in the tomato wilt fungus Fusarium oxysporum f. sp. lycopersici. Journal of Agricultural Research 70: 303–313. [Google Scholar]
- Aloi C, Baayen RP. 1993. Examination of the relationships between vegetative compatibility groups and races in Fusarium oxysporum f. sp. dianthi. Plant Pathology 42: 839–850. [Google Scholar]
- Altinok HH. 2013. Fusarium species isolated from common weeds in eggplants fields and symptomless hosts of Fusarium oxysporum f. sp. melongenae in Turkey. Journal of Phytopathology 161: 335–340. [Google Scholar]
- Altinok HH, Can C. 2010. Characterization of Fusarium oxysporum f. sp. melongenae isolates from eggplant in Turkey by pathogenicity, VCG and RAPD analysis. Phytoparasitica 38: 149–157. [Google Scholar]
- Altinok HH, Can C, Çolak H. 2013. Vegetative compatibility, pathogenicity and virulence diversity of Fusarium oxysporum f. sp. melongenae recovered from eggplant. Journal of Phytopathology 161: 651–660. [Google Scholar]
- Alvarez GLA. 1945. Studies on coffee root disease in Puerto Rico. Journal of Agriculture of the University of Puerto Rico 29: 1–29. [Google Scholar]
- Alves-Santos FM, Cordeiro-Rodrigues L, Sayagués, et al. 2002a. Pathogenicity and race characterization of Fusarium oxysporum f. sp. phaseoli isolates from Spain and Greece. Plant Pathology 51: 605–611. [Google Scholar]
- Alves-Santos FM, Ramos B, García-Sánchez MA, et al. 2002b. A DNA-based procedure for in planta detection of Fusarium oxysporum f. sp. phaseoli. Phytopathology 92: 237–244. [DOI] [PubMed] [Google Scholar]
- Aoki T, O’Donnell K, Geiser DM. 2014. Systematics of key phytopathogenic Fusarium species: current status and future challenges. Journal of General Plant Pathology 80: 189–201. [Google Scholar]
- Arie T, Kaneko I, Yoshida T, et al. 2000. Mating-type genes from asexual phytopathogenic ascomycetes Fusarium oxysporum and Alternaria alternata. Molecular Plant-Microbe Interactions 13: 1330–1339. [DOI] [PubMed] [Google Scholar]
- Armstrong GM. 1954. Alfalfa – a common host for the wilt fusaria from alfalfa, cotton, and cassia. Plant Disease Reporter 38: 221–222. [Google Scholar]
- Armstrong GM, Armstrong JK. 1950. Biological races of the Fusarium causing wilt of cowpea and soybeans. Phytopathology 40: 181–193. [Google Scholar]
- Armstrong GM, Armstrong JK. 1952. Physiologic races of the fusaria causing wilts of the Cruciferae. Phytopathology 42: 255–257. [Google Scholar]
- Armstrong GM, Armstrong JK. 1953. Physiologic races of the wilt fusaria from cabbage, radish, and stock. Phytopathology 43: 465. [Google Scholar]
- Armstrong GM, Armstrong JK. 1958a. A race of the cotton wilt Fusarium causing wilt of Yelredo soybean and flue-cured tobacco. Plant Disease Reporter 42: 147–151. [Google Scholar]
- Armstrong GM, Armstrong JK. 1958b. The Fusarium wilt complex as related to the sweet potato. Plant Disease Reporter 42: 1319–1329. [Google Scholar]
- Armstrong GM, Armstrong JK. 1960. American, Egyptian, and Indian cotton-wilt fusaria; their pathogenicity and relationship to other wilt fusaria. U.S. Department of Agriculture Technical Bulletin 1219: 1–19. [Google Scholar]
- Armstrong GM, Armstrong JK. 1964. Lupinis species – common hosts for wilt fusaria from alfalfa bean, Cassia, cowpea, lupine, and U.S. cotton. Phytopathology 54: 1232–1235. [Google Scholar]
- Armstrong GM, Armstrong JK. 1965. A wilt of soybean caused by a new form of Fusarium oxysporum. Phytopathology 55: 237–239. [Google Scholar]
- Armstrong GM, Armstrong JK. 1966. Races of Fusarium oxysporum f. sp. conglutinans; race 4, new race; and a new host for race 1, Lychnis chalcedonica. Phytopathology 56: 525–530. [Google Scholar]
- Armstrong GM, Armstrong JK. 1968. Formae speciales and races of Fusarium oxysporum causing a tracheomycosis in the syndrome of disease. Phytopathology 58: 1242–1246. [Google Scholar]
- Armstrong GM, Armstrong JK. 1971. Races of the aster-wilt Fusarium. Phytopathology 61: 820–824. [Google Scholar]
- Armstrong GM, Armstrong JK. 1974. Races of Fusarium oxysporum f. sp. pisi, causal agents of wilt of pea. Phytopathology 64: 849–857. [Google Scholar]
- Armstrong GM, Armstrong JK. 1976. Common hosts for Fusarium oxysporum formae speciales spinaciae and betae. Phytopathology 66: 542–545. [Google Scholar]
- Armstrong GM, Armstrong JK. 1978a. A new race (race 6) of the cotton-wilt Fusarium from Brazil. Plant Disease Reporter 62: 421–423. [Google Scholar]
- Armstrong GM, Armstrong JK. 1978b. Formae speciales and races of Fusarium oxysporum causing wilts of the Cucurbitaceae. Phytopathology 68: 19–28. [Google Scholar]
- Armstrong GM, Armstrong JK. 1980. Cowpea wilt Fusarium oxysporum f. sp. tracheiphilum race 1 from Nigeria. Plant Disease 64: 954–955. [Google Scholar]
- Armstrong GM, Armstrong JK. 1981. Formae speciales and races in Fusarium oxysporum causing wilting diseases. In: Nelson PE, Toussoun TA, Cook RJ. (eds), Fusarium: diseases, biology, and taxonomy: 391–399. The Pennsylvania State University Press, Pennsylvania State University, USA. [Google Scholar]
- Armstrong GM, Armstrong JK, Billington RV. 1975. Fusarium oxysporum forma specialis voandzeiae, a new form species causing wilt of Bambarra groundnut. Mycologia 67: 709–714. [PubMed] [Google Scholar]
- Armstrong GM, Armstrong JK, Littrell RH. 1970. Wilt of chrysanthemum caused by Fusarium oxysporum f. sp. chrysanthemi, forma specialis nov. Phytopathology 60: 496–498. [Google Scholar]
- Armstrong GM, Armstrong JK, Netzer D. 1978. Pathogenic races of the cucumber-wilt Fusarium. Plant Disease Reporter 62: 824–828. [Google Scholar]
- Arya HC, Jain GL. 1962. Fusarium wilt of Eucalyptus. Phytopathology 52: 638–642. [Google Scholar]
- Assigbetse KB, Fernandez D, Dubois MP, et al. 1994. Differentiation of Fusarium oxysporum f. sp. vasinfectum races on cotton by random amplified polymorphic DNA (RAPD) analysis. Phytopathology 84: 622–626. [Google Scholar]
- Atkinson GF. 1892. Some diseases of cotton. Bulletin of the Alabama Agricultural Experiment Station 41: 1–65. [Google Scholar]
- Baayen RP, Elgersma DM, Demmink JF, et al. 1988. Differences in pathogenesis observed among susceptible interactions of carnation with four races of Fusarium oxysporum f. sp. dianthi. Netherlands Journal of Plant Pathology 94: 81–94. [Google Scholar]
- Baayen RP, Förch MG, Waalwijk C, et al. 1998. Pathogenic, genetic and molecular characterisation of Fusarium oxysporum f. sp. lilii. European Journal of Plant Pathology 104: 887–894. [Google Scholar]
- Baayen RP, Kleijn J. 1989. The Elegans fusaria causing wilt disease of carnation. II. Distinction of vegetative compatibility groups. Netherlands Journal of Plant Pathology 95: 185–194. [Google Scholar]
- Baayen RP, O’Donnell K, Bonants PJM, et al. 2000. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 90: 891–900. [DOI] [PubMed] [Google Scholar]
- Baayen RP, Van Dreven F, Krijger MC, et al. 1997. Genetic diversity in Fusarium oxysporum f. sp. dianthi and Fusarium redolens f. sp. dianthi. European Journal of Plant Pathology 103: 395–408. [Google Scholar]
- Baker KF. 1948. Fusarium wilt of garden stock (Mattiola incana). Phytopathology 38: 399–403. [Google Scholar]
- Balmas V, Scherm B, Di Primo P, et al. 2005. Molecular characterisation of vegetative compatibility groups in Fusarium oxysporum f. sp. radicis-lycopersici and f. sp. lycopersici by random amplification of polymorphic DNA and microsatellite-primed PCR. European Journal of Plant Pathology 111: 1–8. [Google Scholar]
- Bao JR, Fravel DR, O’Neill, et al. 2002. Genetic analysis of pathogenic and nonpathogenic Fusarium oxysporum from tomato plants. Canadian Journal of Botany 80: 271–279. [Google Scholar]
- Barton E, Borman A, Johnson E, et al. 2016. Pseudo-outbreak of Fusarium oxysporum associated with bronchoscopy. Journal of Hospital Infection 94: 197–198. [DOI] [PubMed] [Google Scholar]
- Barve MP, Haware MP, Sainani MN, et al. 2001. Potential of microsatellites to distinguish four races of Fusarium oxysporum f. sp. ciceri prevalent in India. Theoretical and Applied Genetics 102: 138–147. [Google Scholar]
- Basirnia T, Banihashemi ZAD. 2005. Vegetative compatibility grouping in Fusarium oxysporum f. sp. sesami, the causal agent of sesame yellows and wilt in Fars province. Iranian Journal of Plant Pathology 41: 243–255. [Google Scholar]
- Bayraktar H, Dolar FS, Maden S. 2008. Use of RAPD and ISSR markers in detection of genetic variation and population structure among Fusarium oxysporum f. sp. ciceris isolates on chickpea in Turkey. Journal of Phytopathology 156: 146–154. [Google Scholar]
- Bayraktar H, Türkkan M, Dolar FS. 2010. Characterization of Fusarium oxysporum f. sp. cepae from onion in Turkey based on vegetative compatibility and rDNA RFLP analysis. Journal of Phytopathology 158: 691–697. [Google Scholar]
- Baysal Ö, Karaaslan Ç, Siragusa M, et al. 2013. Molecular markers reflect differentiation of Fusarium oxysporum forma speciales on tomato and forma on eggplant. Biochemical Systematics and Ecology 47: 139–147. [Google Scholar]
- Baysal Ö, Siragusa M, Gümrükcü E, et al. 2010. Molecular characterization of Fusarium oxysporum f. melongenae by ISSR and RAPD markers on eggplant. Biochemical Genetics 48: 524–537. [DOI] [PubMed] [Google Scholar]
- Beach WS. 1918. The Fusarium wilt of china aster. Annual Report of the Michigan Academy of Science 29: 281–308. [Google Scholar]
- Belabid L, Baum M, Fortas Z, et al. 2004. Pathogenic and genetic characterization of Algerian isolates of Fusarium oxysporum f. sp. lentis by RAPD and AFLP analysis. African Journal of Biotechnology 3: 25–31. [Google Scholar]
- Belabid L, Fortas Z. 2002. Virulence and vegetative compatibility of Algerian isolates of Fusarium oxysporum f. sp. lentis. Phytopathologia Mediterranea 41: 179–187. [Google Scholar]
- Bell AA, Wheeler MH, Liu J, et al. 2003. United States Department of Agriculture-Agricultural Research Service studies on polyketide toxins of Fusarium oxysporum f. sp. vasinfectum: Potential targets for disease control. Pest Management Science 59: 736–747. [DOI] [PubMed] [Google Scholar]
- Bennett RS, Hutmacher RB, Davis RM. 2008. Seed transmission of Fusarium oxysporum f. sp. vasinfectum race 4 in California. Journal of Cotton Science 12: 160–164. [Google Scholar]
- Bennett RS, Scott TZ, Lawrence KS, et al. 2013. Sequence characterization of race-4-like isolates of Fusarium oxysporum from Alabama and Mississippi. Journal of Cotton Science 17: 125–130. [Google Scholar]
- Bergstrom GC, Kalb DW. 1995. Fusarium oxysporum f. sp. loti: A specific wilt pathogen of birdsfoot trefoil in New York. Phytopathology 85: 1555. [Google Scholar]
- Bertetti D, Ortu G, Gullino ML, et al. 2014. Contamination of seeds of Iceland poppy (Papaver nudicaule L.) by Fusarium oxysporum. Phytoparasitica 43: 189–196. [Google Scholar]
- Bertetti D, Ortu G, Gullino ML, et al. 2017. Identification of Fusarium oxysporum f. sp. opuntiarum on new hosts of the Cactaceae and Euphorbiaceae families. Journal of Plant Pathology 99: 1–21. [Google Scholar]
- Bertoldo C, Gilardi G, Spadaro D, et al. 2015. Genetic diversity and virulence of Italian strains of Fusarium oxysporum isolated from Eustoma grandiflorum. European Journal of Plant Pathology 141: 83–97. [Google Scholar]
- Bhide VP, Uppal BN. 1948. A new Fusarium disease of lang (Lathyrus sativus). Phytopathology 38: 560–567. [Google Scholar]
- Bilaǐ VI. 1955. The Fusaria (biology and systematics). National Academy of Sciences of Ukraine SSR, Kiev, USSR. [Google Scholar]
- Bilju VC, Fokkens L, Houterman PM, et al. 2017. Multiple evolutionary trajectories have led to the emergence of races in Fusarium oxysporum f. sp. lycopersici. Applied and Environmental Microbiology 83: e02548–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LL, Green SK, Hartman GL, et al. 1993. Enfermedades del Chile. Una Guía de Campo. Ed. Centro Asiatico de Investigación y Desarrollo Vegetal (AVRDC). [Google Scholar]
- Blok WJ, Bollen GJ. 1997. Host specificity and vegetative compatibility of Dutch isolates of Fusarium oxysporum f. sp. asparagi. Canadian Journal of Botany 75: 383–393. [Google Scholar]
- Boerema GH, Hamers MEC. 1989. Check-list for scientific names of common parasitic fungi. Series 3b: Fungi on bulbs: Amaryllidaceae and Iridaceae. Netherlands Journal of Plant Pathology 95 (suppl. 3): 1–32. [Google Scholar]
- Bogale M, Wingfield BD, Wingfield MJ, et al. 2007. Species-specific primers for Fusarium redolens and a PCR-RFLP technique to distinguish among three clades of Fusarium oxysporum. FEMS Microbiology Letters 271: 27–32. [DOI] [PubMed] [Google Scholar]
- Bolley HL. 1901. A preliminary note on the cause of flax-sick soil. Fusarium lini sp. nov. Proceedings of the Annual Meeting of the Society for the Promotion of Agricultural Science 22: 42–46. [Google Scholar]
- Bolton AT, Nuttall VW, Lyall LH. 1966. A new race of Fusarium oxysporum f. sp. pisi. Canadian Journal of Plant Science 46: 343–347. [Google Scholar]
- Booth C. 1971. The genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey, United Kingdom. [Google Scholar]
- Booth C. 1975. The present status of Fusarium taxonomy. Annual Review of Phytopathology 13: 83–93. [Google Scholar]
- Bosland PW, Williams PH. 1987. An evaluation of Fusarium oxysporum from crucifers based on pathogenicity, isozyme polymorphism, vegetative compatibility, and geographic origin. Canadian Journal of Botany 65: 2067–2073. [Google Scholar]
- Brandes EW. 1919. Banana wilt. Phytopathology 9: 339–390. [Google Scholar]
- Brankovics B, Van Dam P, Rep M, et al. 2017. Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex. BMC Genomics 18: 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Phillippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugnicourt F. 1939. Les Fusarium et Cylindrocarpon de l’Indochine. Encyclopédie Mycologique 11: 1–206. [Google Scholar]
- Buxton EW. 1955. The taxonomy and variation in culture of Fusarium oxysporum from gladiolus. Transactions of the British Mycological Society 38: 202–212. [Google Scholar]
- Cai G, Gale R, Schneider RW, et al. 2003. Origin of race 3 of Fusarium oxysporum f. sp. lycopersici at a single site in California. Phytopathology 93: 1014–1022. [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Carlesse F, Amaral A-PC, Gonçalves SS, et al. 2017. Outbreak of Fusarium oxysporum infections in children with cancer: an experience with 7 episodes of catheter-related fungemia. Antimicrobial Resistance and Infection Control 6: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Rep M, Wang B, et al. 2011. Variation in potential effector genes distinguishing Australian and non-Australian isolates of the cotton wilt pathogen Fusarium oxysporum f. sp. vasinfectum. Plant Pathology 60: 232–243. [Google Scholar]
- Chang DC, Grant GB, O’Donnell K, et al. 2006. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA 296: 953–963. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Rai JN. 1974. Fusarium wilt of Eruca vesicaria – observation on comparative pathogenicity of some strains of Fusarium oxysporum. Indian Phytopathology 28: 309–311. [Google Scholar]
- Chen Q, Ji X, Sun W. 1985. Identification of races of cotton wilt Fusarium in China. Agricultural Sciences in China 6: 1–6. [Google Scholar]
- Chen WQ, Swart WJ. 2001. Genetic variation among Fusarium oxysporum isolates associated with root rot of Amaranthus hybridus in South Africa. Plant Disease 85: 1076–1080. [DOI] [PubMed] [Google Scholar]
- Chen ZD, Huang RK, Li QQ, et al. 2015. Development of pathogenicity and AFLP to characterize Fusarium oxysporum f. sp. momordicae isolates from bitter gourd in China. Journal of Phytopathology 163: 202–211. [Google Scholar]
- Chiocchetti A, Ghignone S, Minuto A, et al. 1999. Identification of Fusarium oxysporum f. sp. basilici isolated from soil, basil seed, and plants by RAPD analysis. Plant Disease 83: 576–581. [DOI] [PubMed] [Google Scholar]
- Chiocchetti A, Sciaudone L, Durando F, et al. 2001. PCR detection of Fusarium oxysporum f. sp. basilici on basil. Plant Disease 85: 607–611. [DOI] [PubMed] [Google Scholar]
- Cianchetta AN, Allen TW, Hutmacher RB, et al. 2015. Survey of Fusarium oxysporum f. sp. vasinfectum in the United States. Journal of Cotton Science 19: 328–336. [Google Scholar]
- Coddington A, Matthews PM, Cullis C, et al. 1987. Restriction digest patterns of total DNA from different races of Fusarium oxysporum f. sp. pisi – an improved method for race classification. Journal of Phytopathology 118: 9–20. [Google Scholar]
- Cohen SI. 1946. A wilt and root rot of Asparagus officinalis L. var. altilis L. Phytopathology 36: 397–397. [Google Scholar]
- Correll JC. 1991. The relationship between formae speciales, races, and vegetative compatibility groups in Fusarium oxysporum. Phytopathology 81: 1061–1064. [Google Scholar]
- Correll JC, Klittich CJR, Leslie JF. 1987. Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology 77: 1640–1646. [Google Scholar]
- Correll JC, Puhalla JE, Schneider RW. 1986. Identification of Fusarium oxysporum f. sp. apii on the basis of colony size, virulence, and vegetative compatibility. Phytopathology 76: 396–400. [Google Scholar]
- Covey PA, Kuwitzky B, Hanson M, et al. 2014. Multilocus analysis using putative fungal effectors to describe a population of Fusarium oxysporum from sugar beet. Phytopathology 104: 886–896. [DOI] [PubMed] [Google Scholar]
- Crall JM. 1963. Physiological specialization in Fusarium oxysporum f. sp. niveum. Phytopathology 53: 873. [Google Scholar]
- Cramer RA, Byrne PF, Brick MA, et al. 2003. Characterization of Fusarium oxysporum isolates from common bean and sugar beet using pathogenicity assays and random-amplified polymorphic DNA markers. Journal of Phytopathology 151: 352–360. [Google Scholar]
- Crous PW, Groenewald JZ, Risède JM, et al. 2004. Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. 2009. Fungal Biodiversity. CBS Laboratory manual Series. Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands. [Google Scholar]
- Crowhurst RN, King FY, Hawthorne BT, et al. 1995. RAPD characterization of Fusarium oxysporum associated with wilt of angsana (Pterocarpus indicus) in Singapore. Mycological Research 99: 14–18. [Google Scholar]
- Crutcher FK, Doan HK, Bell AA, et al. 2016. Evaluation of methods to detect the cotton wilt pathogen Fusarium oxysporum f. sp. vasinfectum race 4. European Journal of Plant Pathology 144: 225–230. [Google Scholar]
- Cutuli MT, Gibello A, Rodriquez-Bertos A, et al. 2015. Skin and subcutaneous mycoses in tilapia (Oreochromis niloticus) caused by Fusarium oxysporum in coinfection with Aeromonas hydrophila. Medical Mycology Case Reports 9: 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czislowski E, Fraser-Smith S, Zander M, et al. 2017. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Molecular Plant Pathology 19: 1155–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva FP, Vechiato MH, Harakava R. 2014. EF-1α gene and IGS rDNA sequencing of Fusarium oxysporum f. sp. vasinfectum and F. oxysporum f. sp. phaseoli reveals polyphyletic origins of strains. Tropical Plant Pathology 39: 64–73. [Google Scholar]
- Datta S, Choudhary RG, Shamin M, et al. 2011. Molecular diversity in Indian isolates of Fusarium oxysporum f. sp. lentis inciting wilt disease in lentil (Lens culinaris Medik). African Journal of Biotechnology 10: 7314–7323. [Google Scholar]
- Davis RD, Moore NY, Kochman JK. 1996. Characterisation of a population of Fusarium oxysporum f. sp. vasinfectum causing wilt of cotton in Australia. Australian Journal of Agricultural Research 47: 1143–1156. [Google Scholar]
- De Haan LAM, Numansen A, Roebroeck EJA, et al. 2000. PCR detection of Fusarium oxysporum f. sp. gladioli race 1, causal agent of Gladiolus yellows disease, from infected corms. Plant Pathology 49: 89–100. [Google Scholar]
- De Sousa MV, Machado J da C, Simmons HE, et al. 2015. Real-time quantitative PCR assays for the rapid detection and quantification of Fusarium oxysporum f. sp. phaseoli in Phaseolus vulgaris (common bean) seeds. Plant Pathology 64: 478–488. [Google Scholar]
- De Vega-Bartol JJ, Martín-Dominguez R, Ramos B, et al. 2011. New virulence groups in Fusarium oxysporum f. sp. phaseoli: The expression of the gene coding for the transcription factor ftf1 correlates with virulence. Phytopathology 101: 470–479. [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, et al. 2012. The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton FC, Stevenson JA, Cummins GB. 1962. Formae speciales and the Code. Taxon 11: 70–71. [Google Scholar]
- Demers JE, Garzón CD, Jiménez-Gasco MM. 2014. Striking genetic similarity between races of Fusarium oxysporum f. sp. ciceris confirms a monophyletic origin and clonal evolution of the chickpea vascular wilt pathogen. European Journal of Plant Pathology 139: 309–324. [Google Scholar]
- Desjardins AE. 2006. Fusarium mycotoxins chemistry, genetics and biology. APS Press, St. Paul, USA. [Google Scholar]
- Di Pietro A, Madrid MP, Caracuel Z, et al. 2003. Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Molecular Plant Pathology 4: 315–325. [DOI] [PubMed] [Google Scholar]
- Di Primo P, Cappelli C, Katan T. 2002. Vegetative compatibility groupings of Fusarium oxysporum f. sp. gladioli from saffron. European Journal of Plant Pathology 108: 869–875. [Google Scholar]
- Di Primo P, Cartia G, Katan T. 2001. Vegetative compatibility and heterokaryon in Fusarium oxysporum f. sp. radicis-lycopersici from Italy. Plant Pathology 50: 371–382. [Google Scholar]
- Dita MA, Waalwijk C, Buddenhagen IW, et al. 2010. A molecular diagnostic for tropical race 4 of the banana fusarium wilt pathogen. Plant Pathology 59: 348–357. [Google Scholar]
- Doan HK, Zhang S, Davis RM. 2014. Development and evaluation of AmplifyRP Acceler8 diagnostic assay for the detection of Fusarium oxysporum f. sp. vasinfectum race 4 in cotton. Plant Health Progress. doi: 10.1094/PHP-RS-13-0115. [DOI] [Google Scholar]
- Dong Z, Hsiang T, Luo M, et al. 2017. Draft genome sequence of an isolate of Fusarium oxysporum f. sp. melongenae, the causal agent of Fusarium wilt of eggplant. Genome Announcements 5: e01597–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos Silva A, De Oliveira EJ, Haddad F, et al. 2013. Molecular fingerprinting of Fusarium oxysporum f. sp. passiflorae isolates using AFLP markers. Scientia Agricola 70: 108–115. [Google Scholar]
- Dubey SC, Priyanka K, Singh V, et al. 2012. Race profiling and molecular diversity analysis of Fusarium oxysporum f. sp. ciceris causing wilt of chickpea. Journal of Phytopathology 160: 576–587. [Google Scholar]
- Dubey SC, Singh SR. 2008. Virulence analysis and oligonucleotide fingerprinting to detect diversity among Indian isolates of Fusarium oxysporum f. sp. ciceris causing chickpea wilt. Mycopathologia 165: 389–406. [DOI] [PubMed] [Google Scholar]
- Dzidzariya OM. 1968. Measures for the control of fusariosis of East Indian basil [in Russian]. (Translated in Review of Applied Mycology 48: 882 (1969).) [Google Scholar]
- Edel-Hermann V, Sautour M, Gautheron N, et al. 2016. A clonal lineage of Fusarium oxysporum circulates in the tap water of different French hospitals. Applied and Environmental Microbiology 82: 6483–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egamberdiev SS, Salahutdinov IB, Abdullaev AA, et al. 2014. Detection of Fusarium oxysporum f. sp. vasinfectum race 3 by single-base extension method and allele-specific polymerase chain reaction. Canadian Journal of Plant Pathology 36: 216–223. [Google Scholar]
- Egamberdiev SS, Ulloa M, Saha S, et al. 2013. Molecular characterization of Uzbekistan isolates of Fusarium oxysporum f. sp. vasinfectum. Journal of Plant Science & Molecular Breeding. doi: 10.7243/2050-2389-2-3. [DOI] [Google Scholar]
- Elias KS, Schneider RW. 1991. Vegetative compatibility groups in Fusarium oxysporum f. sp. lycopersici. Phytopathology 81: 159–162. [Google Scholar]
- Elias KS, Schneider RW. 1992. Genetic diversity within and among races and vegetative compatibility groups of Fusarium oxysporum f. sp. lycopersici as determined by isozyme analysis. Phytopathology 82: 1421–1427. [Google Scholar]
- Elias KS, Zamir D, Lichtman-Pleban T, et al. 1993. Population structure of Fusarium oxysporum f. sp. lycopersici: restriction fragment length polymorphisms provide genetic evidence that vegetative compatibility group is an indicator of evolutionary origin. Molecular Plant-Microbe Interaction 6: 565–572. [Google Scholar]
- Elliott ML, Des Jardin EA, Harmon CL, et al. 2017. Confirmation of Fusarium wilt caused by Fusarium oxysporum f. sp. palmarum on × Butyagrus nabonnandii (mule palm) in Florida. Plant Disease 101: 381. [Google Scholar]
- Elliott ML, Des Jardin EA, O’Donnell K, et al. 2010. Fusarium oxysporum f. sp. palmarum, a novel forma specialis causing a lethal disease of Syagrus romanzoffiana and Washingtonia robusta in Florida. Plant Disease 94: 31–38. [DOI] [PubMed] [Google Scholar]
- Elmer WH, Stephens CT. 1989. Classification of Fusarium oxysporum f. sp. asparagi into vegetatively compatible groups. Phytopathology 79: 88–93. [Google Scholar]
- Elmer WH, Wick RL, Haviland P. 1994. Vegetative compatibility among Fusarium oxysporum f. sp. basilicum isolates recovered from basil seed and infected plants. Plant Disease 78: 789–791. [Google Scholar]
- Elzein A, Brändle F, Cadisch G, et al. 2008. Fusarium oxysporum strains as potential Striga mycoherbicides: molecular characterization and evidence for a new forma specialis. The Open Mycology Journal 2: 89–93. [Google Scholar]
- Elzein A, Kroschel J. 2006. Host range studies of Fusarium oxysporum Foxy 2: an evidence for a new forma specialis and its implications for Striga control. Journal of Plant Diseases and Protection 20: 875–887. [Google Scholar]
- Enya J, Togawa M, Takeuchi T, et al. 2008. Biological and phylogenetic characterization of Fusarium oxysporum complex, which causes yellows on Brassica spp., and proposal of Fusarium oxysporum f. sp. rapae, a novel forma specialis pathogenic on Brassica rapa L. in Japan. Phytopathology 98: 475–483. [DOI] [PubMed] [Google Scholar]
- Epstein L, Kaur S, Chang PL, et al. 2017. Races of the celery pathogen Fusarium oxysporum f. sp. apii are polyphyletic. Phytopathology 107: 463–473. [DOI] [PubMed] [Google Scholar]
- Erwin DC. 1958. Fusarium lateritium f. ciceri incitant of Fusarium wilt of Cicer arietinum. Phytopathology 48: 498–501. [Google Scholar]
- Fang X, Jost R, Finnegan PM, et al. 2013. Comparative proteome analysis of the strawberry-Fusarium oxysporum f. sp. fragariae pathosystem reveals early activation of defence responses as a crucial determinant of host resistance. Journal of Proteome 12: 1772–1788. [DOI] [PubMed] [Google Scholar]
- Feather TV, Ohr HD, Munnecke DE. 1979. Wilt and dieback of Canary Island palm in California. California Agriculture 33: 19–20. [Google Scholar]
- Fernandez D, Assigbetse K, Dubois MP, et al. 1994. Molecular characterization of races and vegetative compatibility groups in Fusarium oxysporum f. sp. vasinfectum. Applied and Environmental Microbiology 60: 4039–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D, Ouinten M, Tantaoui A, et al. 1998. Fot 1 insertions in the Fusarium oxysporum f. sp. albedinis genome provide diagnostic PCR targets for detection of the date palm pathogen. Applied and Environmental Microbiology 64: 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher NL, Burgess LW, Toussoun TA, et al. 1982. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72: 151–153. [Google Scholar]
- Flood J. 2006. A review of Fusarium wilt of oil palm caused by Fusarium oxysporum f. sp. elaeidis. Phytopathology 96: 660–662. [DOI] [PubMed] [Google Scholar]
- Foster V. 1955. Fusarium wilt of cattleyas. Phytopathology 45: 599–602. [Google Scholar]
- Fourie G, Steenkamp ET, Ploetz RC, et al. 2011. Current status of the taxonomic position of Fusarium oxysporum formae specialis cubense within the Fusarium oxysporum complex. Infection, Genetics and Evolution 11: 533–542. [DOI] [PubMed] [Google Scholar]
- Fujinaga M, Ogiso H, Shinohara H, et al. 2005. Phylogenetic relationships between the lettuce root rot pathogen Fusarium oxysporum f. sp. lactucae races 1, 2, and 3 based on the sequence of the intergenic spacer region of its ribosomal DNA. Journal of General Plant Pathology 71: 402–407. [Google Scholar]
- Fujinaga M, Ogiso H, Tsuchiya N, et al. 2001. Physiological specialization of Fusarium oxysporum f. sp. lactucae, a causal organism of Fusarium root rot of crisp head lettuce in Japan. Journal of General Plant Pathology 67: 205–206. [Google Scholar]
- Fujinaga M, Ogiso H, Tsuchiya N, et al. 2003. Race 3, a new race of Fusarium oxysporum f. sp. lactucae determined by a differential system with commercial cultivars. Journal of General Plant Pathology 69: 23–28. [Google Scholar]
- Fujinaga M, Yamagishi N, Ishiyama Y, et al. 2014. PCR-based race differentiation of Fusarium oxysporum f. sp. lactucae. Annual Report of the Kanto-Tosan Plant Protection Society 61: 47–49. [Google Scholar]
- Gabe HL. 1975. Standardization of nomenclature for pathogenic races of Fusarium oxysporum f. sp. lycopersici. Transactions of the British Mycological Society 64: 156–159. [Google Scholar]
- Galván GA, Koning-Boucoiran CFS, Koopman WJM, et al. 2008. Genetic variation among Fusarium isolates from onion, and resistance to Fusarium basal rot in related Allium species. European Journal of Plant Pathology 121: 499–512. [Google Scholar]
- García-Pedrasjas MD, Bainbridge BW, Heale JB, et al. 1999. A simple PCR-based method for the detection of the chickpea-wilt pathogen Fusarium oxysporum f.sp. ciceris in artificial and natural soils. European Journal of Plant Pathology 105: 251–259. [Google Scholar]
- Gardner DE. 1980. Acacia koa seedling wilt caused by Fusarium oxysporum f. sp. koae f. sp. nov. Phytopathology 70: 594–597. [Google Scholar]
- Garibaldi A. 1975. Race differentiation in Fusarium oxysporum f. sp. dianthi (Prill et Del.) Snyd. et Hans. First contribution. Mededelingen Faculteit Landbouwwetenschappen Rijksuniversiteit Gent 40: 531–537. [Google Scholar]
- Garibaldi A. 1977. Race differentiation in Fusarium oxysporum f. sp. dianthi and varietal susceptibility. Acta Horticulturae 71: 97–101. [Google Scholar]
- Garibaldi A. 1983. Resistenza di cultivar di garofano nei confronti di otto patotipi di Fusarium oxysporum f. sp. dianthi (Prill. et Del.) Snyd. et Hans. Rivista della Ortoflorofrutticoltura Italiana 67: 261–270. [Google Scholar]
- Garibaldi A, Gullino ML. 1985. Wilt of Ranunculus asiaticus caused by Fusarium oxysporum f. sp. ranunculi, forma specialis nova. Phytopathologia Mediterranea 24: 213–214. [Google Scholar]
- Gawehns F, Houterman PM, Ichou FA, et al. 2014. The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Molecular Plant-Microbe Interactions 27: 336–348. [DOI] [PubMed] [Google Scholar]
- Gerdemann JW, Finley AM. 1951. The pathogenicity of races 1 and 2 of Fusarium oxysporum f. lycopersici. Phytopathology 41: 238–244. [Google Scholar]
- Geiser DM, Aoki T, Bacon CW, et al. 2013. One fungus, one name: Defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 103: 400–408. [DOI] [PubMed] [Google Scholar]
- Gerlach W. 1954. Untersuchungen über die Welkekrankheit des Alpenveilchens. Phytopathologische Zeitschrift 22: 125–176. [Google Scholar]
- Gerlach W, Nirenberg H. 1982. The genus Fusarium – a pictorial atlas. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 209: 1–406. [Google Scholar]
- Gerlagh M, Blok WJ. 1988. Fusarium oxysporum f. sp. cucurbitacearum n.f. embracing all formae speciales of F. oxysporum attacking cucurbitaceous crops. Netherlands Journal of Plant Pathology 94: 17–31. [Google Scholar]
- Gerlagh M, Ester A. 1985. Fusarium oxysporum f. sp. benincasae, a new adaptation of Fusarium oxysporum to a cucurbitaceous crop. Mededelingen van de Faculteit Landbouwwetenschappen 37: 1048. [Google Scholar]
- Gherbawy YAMH. 1999. RAPD profile analysis of isolates belonging to different formae speciales of Fusarium oxysporum. Cytologia 64: 269–276. [Google Scholar]
- Ghosh R, Nagavardhini A, Sengupta A, et al. 2015. Development of loop-mediated isothermal amplification (LAMP) assay for rapid detection of Fusarium oxysporum f. sp. ciceris – wilt pathogen of chickpea. BMC Research Notes 8: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbrecht M, McCarthy M, Elliott ML, et al. 2013. First report of Fusarium oxysporum f. sp. palmarum in Texas causing Fusarium wilt of Washington robusta. Plant Disease 97: 1511. [DOI] [PubMed] [Google Scholar]
- Gilardi G, Franco Ortega S, Van Rijswick PCJ, et al. 2017. A new race of Fusarium oxysporum f. sp. lactucae of lettuce. Plant Pathology 66: 677–688. [Google Scholar]
- Gordon TR. 2017. Fusarium oxysporum and the Fusarium wilt syndrome. Annual Review of Phytopathology 55: 23–39. [DOI] [PubMed] [Google Scholar]
- Gordon TR, Martyn RD. 1997. The evolutionary biology of Fusarium oxysporum. Annual Review of Phytopathology 35: 111–128. [DOI] [PubMed] [Google Scholar]
- Gordon WL. 1965. Pathogenic strains of Fusarium oxysporum. Canadian Journal of Botany 43: 1309–1318. [Google Scholar]
- Grajal-Martin MJ, Simon CJ, Muehlbauer FJ. 1993. Use of random amplified polymorphic DNA (RAPD) to characterize race 2 of Fusarium oxysporum f. sp. pisi. Phytopathology 83: 612–614. [Google Scholar]
- Groenewald JZ, Nakashima C, Nishikawa J, et al. 2013. Species concepts in Cercospora: spotting the weeds among the roses. Studies in Mycology 75: 115–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarro J. 2013. Fusariosis, a complex infection caused by a high diversity of fungal species refractory to treatment. European Journal of Clinical Microbiology & Infectious Diseases 32: 1491–1500. [DOI] [PubMed] [Google Scholar]
- Guarro J, Gene J. 1995. Opportunistic Fusarial infections in humans. European Journal of Clinical Microbiology & Infectious Diseases 14: 741–754. [DOI] [PubMed] [Google Scholar]
- Gunn LV, Summerell BA. 2002. Differentiation of Fusarium oxysporum isolates from Phoenix canariensis (Canary Island date palm) by vegetative compatibility grouping and molecular analysis. Australasian Plant Pathology 31: 351–358. [Google Scholar]
- Guo Q, Li S, Lu X, et al. 2015. Identification of a new genotype of Fusarium oxysporum f. sp. vasinfectum on cotton in China. Plant Disease 99: 1569–1577. [DOI] [PubMed] [Google Scholar]
- Gupta D, Chowdhry PN, Padhi B. 1982. A new form of Fusarium oxysporum on Sansevieria cylindrica. Indian Phytopathology 35: 695–696. [Google Scholar]
- Gupta M, Jarial K, Vikram A. 2014. Morphological, cultural, pathological and molecular variability among Fusarium oxysporum f. sp. zingiberti isolates. International Journal of Bio-resource and Stress Management 5: 375–380. [Google Scholar]
- Gupta PKS. 1974. Plant pathogenic species of the genus Fusarium in India. Nova Hedwigia 25: 699–717. [Google Scholar]
- Gupta VK. 2012. PCR-RAPD profiling of Fusarium spp. causing guava wilt disease in India. Journal of Environmental Science and Health 47: 315–325. [DOI] [PubMed] [Google Scholar]
- Gurjar G, Barve M, Giri A, et al. 2009. Identification of Indian pathogenic races of Fusarium oxysporum f. sp. ciceris with gene specific, ITS and random markers. Mycologia 101: 484–495. [DOI] [PubMed] [Google Scholar]
- Hadar E, Katan J, Katan T. 1989. The use of nitrate-nonutilizing mutants and a selective medium for studies of pathogenic strains of Fusarium oxysporum. Plant Disease 73: 800–803. [Google Scholar]
- Haglund WA, Kraft JM. 1979. Fusarium oxysporum f. sp. pisi, race 6: Occurrence and distribution. Phytopathology 69: 818–820. [Google Scholar]
- Hannachi I, Poli A, Rezgui S, et al. 2015. Genetic and phenotypic differences of Fusarium oxysporum f. sp. citri isolated from sweet orange and tangerine. European Journal of Plant Pathology 142: 269–280. [Google Scholar]
- Hansen FT, Gardiner DM, Lysøe E, et al. 2015. An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal Genetics and Biology 75: 20–29. [DOI] [PubMed] [Google Scholar]
- Hanzawa J. 1914. Fusarium cepae, ein neuer zweibelpilz Japans, sowie einige andere pilze an zweibelpflanzen. Mykologie Zentralblatt 5: 4–13. [Google Scholar]
- Hare WW. 1953. A new race causing wilt of cowpea. Phytopathology 43: 291. [Google Scholar]
- Hartig R. 1892. Ein neuer Keimlingspilz. Zeitschrift für Naturwissenschaften 1: 432–436. [Google Scholar]
- Harveson RM, Rush CM. 1997. Genetic variation among Fusarium oxysporum isolates from sugar beet as determined by vegetative compatibility. Plant Disease 81: 85–88. [DOI] [PubMed] [Google Scholar]
- Haware MP, Nene YL. 1982. Races of Fusarium oxysporum f. sp. ciceri. Plant Disease 66: 809–810. [Google Scholar]
- Hemo I, Pe’Er J, Polacheck I. 1989. Fusarium oxysporum keratitis. Ophthalmologica 198: 3–7. [DOI] [PubMed] [Google Scholar]
- Henrique FH, Carbonell SAM, Ito MF, et al. 2015. Classification of physiological races of Fusarium oxysporum f. sp. phaseoli in common bean. Bragantia 74: 84–92. [Google Scholar]
- Henry PM, Kirkpatrick SC, Islas CM, et al. 2017. The population of Fusarium oxysporum f. sp. fragariae, cause of Fusarium wilt of strawberry, in California. Plant Disease 101: 550–556. [DOI] [PubMed] [Google Scholar]
- Hibar K, Edel-Herman V, Steinberg C, et al. 2007. Genetic diversity of Fusarium oxysporum populations isolated from tomato plants in Tunisia. Journal of Phytopathology 155: 136–142. [Google Scholar]
- Hill AL, Reeves PA, Larson RL, et al. 2011. Genetic variability among isolates of Fusarium oxysporum from sugar beet. Plant Pathology 60: 496–505. [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. [Google Scholar]
- Hirano Y, Arie T. 2006. PCR-based differentiation of Fusarium oxysporum ff. sp. lycopersici and radicis-lycopersici and races of F. oxysporum f. sp. lycopersici. Journal of General Plant Pathology 72: 273–283. [Google Scholar]
- Hirano Y, Arie T. 2009. Variation and phylogeny of Fusarium oxysporum isolates based on nucleotide sequences of polygalacturonase genes. Microbes and Environments 24: 113–120. [DOI] [PubMed] [Google Scholar]
- Hirooka Y, Matsumoto Y, Ebihara Y, et al. 2008. Occurrence of Fusarium wilt in Japan caused by Fusarium oxysporum f. sp. tanaceti. Japanese Journal of Phytopathology 74: 7–12. [Google Scholar]
- Holmes EA, Bennett RS, Spurgeon DW, et al. 2009. New genotypes of Fusarium oxysporum f. sp. vasinfectum from the southeastern United States. Plant Disease 93: 1298–1304. [DOI] [PubMed] [Google Scholar]
- Honnareddy N, Dubey SC. 2006. Pathogenic and molecular characterization of Indian isolates of Fusarium oxysporum f. sp. ciceris causing chickpea wilt. Current Science 91: 661–666. [Google Scholar]
- Hood JR, Stewart RN. 1957. Factors affecting symptom expression in Fusarium wilt of Dianthus. Phytopathology 47: 173–178. [Google Scholar]
- Huang CH, Roberts PD, Gale LR, et al. 2013. Population structure of Fusarium oxysporum f. sp. radicis-lycopersici in Florida inferred from vegetative compatibility groups and microsatellites. European Journal of Plant Pathology 136: 509–521. [Google Scholar]
- Huang HC, Phillippe LM, Marshall HH, et al. 1992. Wilt of hardy chrysanthemum caused by a new race of Fusarium oxysporum f. sp. chrysanthemi. Plant Pathology 1: 57–61. [Google Scholar]
- Huang L-W-, Wang C-J, Lin Y-S, et al. 2014. Stem rot of jewel orchids caused by a new forma specialis, Fusarium oxysporum f. sp. anoectochili in Taiwan. Plant Pathology 63: 539–547. [Google Scholar]
- Hubbard JC, Gerik JS. 1993. A new wilt disease of lettuce incited by Fusarium oxysporum f. sp. lactucum forma specialis nov. Plant Disease 77: 750–754. [Google Scholar]
- Hungerford CW. 1923. A Fusarium wilt of spinach. Phytopathology 13: 205–209. [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23: 254–267. [DOI] [PubMed] [Google Scholar]
- Ibrahim FM. 1966. A new race of cotton-wilt Fusarium in the Sudan Gezira. Empire Cotton Growing Review 43: 296–299. [Google Scholar]
- Imle EP. 1942. Bulbrot disease of lilies. The Lily Yearbook of the American Horticultural Society: 30–41. [Google Scholar]
- Inami K, Yoshioka C, Hirano Y, et al. 2010. Real-time PCR for differential determination of the tomato wilt fungus, Fusarium oxysporum f. sp. lycopersici, and its races. Journal of General Plant Pathology 76: 116–121. [Google Scholar]
- Jacobson DJ, Gordon TR. 1988. Vegetative compatibility and self-incompatibility within Fusarium oxysporum f. sp. melonis. Phytopathology 78: 668–672. [Google Scholar]
- Jacobson DJ, Gordon TR. 1990a. Further investigations of vegetative compatibility within Fusarium oxysporum f. sp. melonis. Canadian Journal of Botany 68: 1245–1248. [Google Scholar]
- Jacobson DJ, Gordon TR. 1990b. Variability of mitochondrial DNA as an indicator of relationships between populations of Fusarium oxysporum f. sp. melonis. Mycological Research 94: 734–744. [Google Scholar]
- Janardhanan KK, Ganguly D, Husain A. 1964. Fusarium wilt of Rauvolfia serpentina. Current Science 33: 313. [Google Scholar]
- Janson BF. 1951. A new disease of dill. Phytopathology 41: 19. [Google Scholar]
- Jarvis WR, Shoemaker RA. 1978. Taxonomic status of Fusarium oxysporum causing foot and root rot of tomato. Phytopathology 68: 1679–1680. [Google Scholar]
- Jelinski NA, Broz K, Jonkers W, et al. 2017. Effector gene suites in some soil isolates of Fusarium oxysporum are not sufficient predictors of vascular wilt of tomato. Phytopathology 107: 842–851. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Al-Hatmi AMS, Xiang Y, et al. 2016. The concept of ecthyma gangrenosum illustrated by a Fusarium oxysporum infection in an immunocompetent individual. Mycopathologia 181: 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Gasco MM, Jiménez-Díaz RM. 2003. Development of a specific polymerase chain reaction-based assay for the identification of Fusarium oxysporum f. sp. ciceris and its pathogenic races 0, 1A, 5, and 6. Phytopathology 93: 200–209. [DOI] [PubMed] [Google Scholar]
- Jiménez-Gasco MM, Milgroom MG, Jiménez-Díaz RM. 2002. Gene genealogies support Fusarium oxysporum f. sp. ciceris as a monophyletic group. Plant Pathology 51: 72–77. [Google Scholar]
- Jiménez-Gasco MM, Milgroom MG, Jiménez-Díaz RM. 2004a. Stepwise evolution of races in Fusarium oxysporum f. sp. ciceris inferred from fingerprinting with repetitive DNA sequences. Phytopathology 94: 228–235. [DOI] [PubMed] [Google Scholar]
- Jiménez-Gasco MM, Navas-Cortés JA, Jiménez-Díaz RM. 2004b. The Fusarium oxysporum f. sp. ciceris/Cicer arietinum pathosystem: a case study of the evolution of plant-pathogenic fungi into races and pathotypes. International Microbiology 7: 95–104. [PubMed] [Google Scholar]
- Jiménez-Gasco MM, Pérez-Artés E, Jiménez-Díaz RM. 2001. Identification of pathogenic races 0, 1B/C, 5, and 6 of Fusarium oxysporum f. sp. ciceris with random amplified polymorphic DNA (RAPD). European Journal of Plant Pathology 107: 237–248. [Google Scholar]
- Johnson J. 1921. Fusarium-wilt of tobacco. Journal of Agriculture Research 20: 515–535. [Google Scholar]
- Kappelman AJ. 1983. Distribution of races of Fusarium oxysporum f. sp. vasinfectum within the United States. Plant Disease 67: 1229–1231. [Google Scholar]
- Kashiwa T, Inami K, Teraoka T, et al. 2016. Detection of cabbage yellows fungus Fusarium oxysporum f. sp. conglutinans in soil by PCR and real-time PCR. Journal of General Plant Pathology 82: 240–247. [Google Scholar]
- Katan T. 1999. Current status of vegetative compatibility groups in Fusarium oxysporum. Phytoparasitica 27: 51–64. [Google Scholar]
- Katan T, Di Primo P. 1999. Current status of vegetative compatibility groups in Fusarium oxysporum: Supplement (1999). Phytoparasitica 27: 273–277. [Google Scholar]
- Katan T, Katan J. 1988. Vegetative-compatibility grouping of Fusarium oxysporum f. sp. vasinfectum from tissue and the rhizosphere of cotton plants. Phytopathology 78: 852–855. [Google Scholar]
- Katan T, Katan J, Gordon TR, et al. 1994. Physiological races and vegetative compatibility groups of Fusarium oxysporum f. sp. melonis in Israel. Phytopathology 84: 153–157. [Google Scholar]
- Katan T, Zamir D, Sarfatti M, et al. 1991. Vegetative compatibility groups and subgroups in Fusarium oxysporum f. sp. radicis-lycopersici. Phytopathology 81: 255–262. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics: 1–7. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe M, Katsube K, Yoshida T, et al. 2007. Genetic diversity of Fusarium oxysporum f. sp. spinaciae in Japan based on phylogenic analyses of rDNA-IGS and MAT1 sequences. Journal of General Plant Pathology 73: 353–359. [Google Scholar]
- Kawai I, Suzuki H, Kawai K. 1958. On the pathogenicity of wilt Fusarium of the cucurbitaceous plants and their forms. Schizuoka Agricultural Experiment Station Bulletin 3: 49–68. [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A, Alcalá-Jiménez AR, Bainbridge BW, et al. 1994. Use of genetic fingerprinting and random amplified polymorphic DNA to characterize pathotypes of Fusarium oxysporum f. sp. ciceris infecting chickpea. Phytopathology 84: 1293–1298. [Google Scholar]
- Kelly AG, Bainbridge BW, Heale JB, et al. 1998. In planta-polymerase-chain-reaction detection of the wilt-inducing pathotype of Fusarium oxysporum f. sp. ciceris in chickpea (Cicer arietinum L.). Physiological and Molecular Plant Pathology 52: 397–409. [Google Scholar]
- Kendrick JB, Snyder WC. 1942a. Fusarium wilt of radish. Phytopathology 32: 1031–1033. [Google Scholar]
- Kendrick JB, Snyder WC. 1942b. Fusarium yellows of beans. Phytopathology 32: 1010–1014. [Google Scholar]
- Khetan S, Khetan P, Katkar V, et al. 2018. Urinary tract infection due to Fusarium oxysporum in an immunocompetent patient with chronic kidney disease. Journal of Biomedical Research 32: 157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian C, Maire R. 1930. Le Bayoud, maladie du dattier. Bulletin de la Société d’Histoire Naturelle de l’Afrique du Nord 21: 89–101. [Google Scholar]
- Kim DH, Martyn RD, Magill CW. 1993. Mitochondrial DNA (mtDNA) – Relatedness among formae speciales of Fusarium oxysporum in the Cucurbitaceae. Phytopathology 83: 91–97. [Google Scholar]
- Kim H, Hwang SM, Lee JH, et al. 2017. Specific PCR detection of Fusarium oxysporum f. sp. raphani: a causal agent of Fusarium wilt on radish plants. Plant Pathology 65: 133–140. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Choi YK, Min BR. 2001. Variation of the intergenic spacer (IGS) region of ribosomal DNA among Fusarium oxysporum formae speciales. Journal of Microbiology 39: 265–272. [Google Scholar]
- Kim WS, Kim WG, Cho WD, et al. 2002. Wilt of perilla caused by Fusarium spp. Plant Pathology Journal 18: 293–299. [Google Scholar]
- Kim Y, Hutmacher RB, Davis RM. 2005. Characterization of California isolates of Fusarium oxysporum f. sp. vasinfectum. Plant Disease 89: 366–372. [DOI] [PubMed] [Google Scholar]
- Kistler HC. 1997. Genetic diversity in the plant-pathogenic fungus Fusarium oxysporum. Phytopathology 87: 474–479. [DOI] [PubMed] [Google Scholar]
- Kistler HC, Alabouvette C, Baayen RP, et al. 1998. Systematic numbering of vegetative compatibility groups in the plant pathogenic fungus Fusarium oxysporum. Phytopathology 88: 30–32. [DOI] [PubMed] [Google Scholar]
- Kistler HC, Benny U. 1989. The mitochondrial genome of Fusarium oxysporum. Plasmid 22: 86–89. [DOI] [PubMed] [Google Scholar]
- Kistler HC, Bosland PW, Benny U, et al. 1987. Relatedness of strains of Fusarium oxysporum from crucifers measured by examination of mitochondrial and ribosomal DNA. Phytopathology 77: 1289–1293. [Google Scholar]
- Kistler HC, Momol EA, Benny U. 1991. Repetitive genomic sequences for determining relatedness among strains of Fusarium oxysporum. Phytopathology 81: 331–336. [Google Scholar]
- Kitazawa K, Yanagita K. 1984. Adzuki bean wilt caused by Fusarium oxysporum Schl.: Re-occurrence and confirmation of the causal organism. Annals of the Phytopathological Society of Japan 50: 643–645. [Google Scholar]
- Kitazawa K, Yanagita K. 1989. Fusarium oxysporum Schl. F. sp. adzukicola n.f. sp., a wilt fungus of Phaseolus angularis. Annals of the Phytopathological Society of Japan 55: 76–78. [Google Scholar]
- Klisiewicz JM. 1975. Race 4 of Fusarium oxysporum f. sp. carthami. Plant Disease Reporter 59: 712–714. [Google Scholar]
- Klisiewicz JM, Houston BR. 1963. A new form of Fusarium oxysporum. Phytopathology 53: 241. [Google Scholar]
- Klisiewicz JM, Thomas CA. 1970a. Pathogenic races of Fusarium oxysporum f. sp. carthami. Phytopathology 60: 83–84. [Google Scholar]
- Klisiewicz JM, Thomas CA. 1970b. Race differentiation in Fusarium oxysporum f. sp. carthami. Phytopathology 60: 1706. [Google Scholar]
- Kondo T, Chu E, Kageyama K, et al. 2013. Stem canker and wilt of delphinium caused by Fusarium oxysporum f. sp. delphinii in Japan. Journal of General Plant Pathology 79: 370–373. [Google Scholar]
- Koyyappurath S, Atuahiva T, Le Guen R, et al. 2016. Fusarium oxysporum f. sp. radicis-vanillae is the causal agent of root and stem rot of vanilla. Plant Pathology 65: 612–625. [Google Scholar]
- Kraft JM, Haglund WA. 1978. A reappraisal of the race classification of Fusarium oxysporum f. sp. pisi. Phytopathology 68: 273–275. [Google Scholar]
- Kulkarni GS. 1934. Studies in the wilt disease of cotton in the Bombay presidency. Indian Journal of Agricultural Sciences 4: 976–1045. [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RP, Hopkins DL, Martin FN. 1988. Differentiation of strains and pathogenic races of Fusarium oxysporum f. sp. niveum based on vegetative compatibility. Phytopathology 88: 1542. [Google Scholar]
- Larkin RP, Hopkins DL, Martin FN. 1990. Vegetative compatibility within Fusarium oxysporum f. sp. niveum and its relationship to virulence, aggressiveness, and race. Canadian Journal of Microbiology 36: 352–358. [Google Scholar]
- Laskaris T. 1949. Fusarium stem canker and wilt of Delphinium. Phytopathology 39: 913–919. [Google Scholar]
- Laurence MH, Burgess LW, Summerell BA, et al. 2012. High levels of diversity in Fusarium oxysporum from non-cultivated ecosystems in Australia. Fungal Biology 116: 289–297. [DOI] [PubMed] [Google Scholar]
- Laurence MH, Summerell BA, Burgess LW, et al. 2014. Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biology 118: 374–384. [DOI] [PubMed] [Google Scholar]
- Laurence MH, Summerell BA, Liew ECY. 2015. Fusarium oxysporum f. sp. canariensis: evidence for horizontal gene transfer of putative pathogenicity genes. Plant Pathology 64: 1068–1075. [Google Scholar]
- Leach JG, Currence TM. 1938. Fusarium wilt of muskmelons in Minnesota. Minnesota Agricultural Experiment Station Technical Bulletin 129: 1–32. [Google Scholar]
- Lecomte C, Edel-Hermann V, Cannesan M-A, et al. 2016. Fusarium oxysporum f. sp. cyclaminis: underestimated genetic diversity. European Journal of Plant Pathology 145: 421–431. [Google Scholar]
- Leslie JF. 1993. Vegetative compatibility in fungi. Annual Review of Phytopathology 31: 127–151. [DOI] [PubMed] [Google Scholar]
- Leslie JF, Summerell BA. 2006. The Fusarium laboratory manual. Blackwell Publishing, Ames. [Google Scholar]
- Li D, Wang L, Zhang Y, et al. 2012. Pathogenic variation and molecular characterization of Fusarium species isolated from wilted sesame in China. African Journal of Microbiology Research 6: 149–154. [Google Scholar]
- Li E, Ling J, Wang G, et al. 2015. Comparative proteomics analyses of two races of Fusarium oxysporum f. sp. conglutinans that differ in pathogenicity. Scientific Reports 5: 13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Wang G, Xiao J, et al. 2016. A SIX1 homolog in Fusarium oxysporum f. sp. conglutinans is required for full virulence on cabbage. PLoS ONE 11 (3): e0152273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Garibaldi A, Gullino ML. 2010. Molecular detection of Fusarium oxysporum f. sp. chrysanthemi on three host plants: Gerbera, jamesonii, Osteospermum sp. and Argyranthemum frutescens. Journal of Plant Pathology 92: 525–530. [Google Scholar]
- Lievens B, Claes L, Vakalounakis DJ, et al. 2007. A robust identification and detection assay to discriminate the cucumber pathogens Fusarium oxysporum f. sp. cucumerinum and f. sp. radicis-cucumerinum. Environmental Microbiology 9: 2145–2161. [DOI] [PubMed] [Google Scholar]
- Lievens B, Houterman PM, Rep M. 2009a. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiology Letters 300: 201–215. [DOI] [PubMed] [Google Scholar]
- Lievens B, Rep M, Thomma BPHJ. 2008. Recent development in the molecular discrimination of formae speciales of Fusarium oxysporum. Pest Management Science 64: 781–788. [DOI] [PubMed] [Google Scholar]
- Lievens B, Van Baarlen P, Verreth C, et al. 2009b. Evolutionary relationships between Fusarium oxysporum f. sp. lycopersici and Fusarium oxysporum f. sp. radicis-lypersici isolates inferred from mating, type, elongation factor-1α and exopolygalacturonase sequences. Mycological Research 113: 1181–1191. [DOI] [PubMed] [Google Scholar]
- Lin B, Shen H. 2017. Fusarium oxysporum f. sp. cubense. In: Wan F, Jiang M, Zhan A. (eds), Biological invasions and its management in China: 225–236. Springer, Singapore. [Google Scholar]
- Lin Q, Chen Z. 1994. Identification of the causal agent of root rot of Magnolia biloba. Acta Phytopathologica Sinica 24: 312. [Google Scholar]
- Lin YH, Chen KS, Chang JY, et al. 2010. Development of the molecular methods for rapid detection and differentiation of Fusarium oxysporum and F. oxysporum f. sp. niveum in Taiwan. New Biotechnology 27: 409–418. [DOI] [PubMed] [Google Scholar]
- Lin YH, Lai PJ, Chang TH, et al. 2014. Genetic diversity and identification of race 3 of Fusarium oxysporum f. sp. lactucae in Taiwan. European Journal of Plant Pathology 140: 721–733. [Google Scholar]
- Linfield CA. 1993. A rapid serological test for detecting Fusarium oxysporum f. sp. narcissi in Narcissus. Annals of Applied Biology 123: 685–693. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Löffler HJM, Rumine P. 1991. Virulence and vegetative compatibility of Dutch and Italian isolates of Fusarium oxysporum f. sp. lilii. Journal of Phytopathology 132: 12–20. [Google Scholar]
- Logrieco A, Moretti A, Castellá G, et al. 1998. Beauvericin production by Fusarium species. Applied and Environmental Microbiology 64: 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas-Cano T, Boix-Ruiz A, De Cara-García M, et al. 2016. Etiological and epidemiological concerns about pepper root and lower stem rot caused by Fusarium oxysporum f. sp. radicis-capsici f. sp. nova. Phytoparasitica 44: 283–293. [Google Scholar]
- Lomas-Cano T, Palmero-Liamas D, De Cara M, et al. 2014. First report of Fusarium oxysporum on sweet pepper seedlings in Almería, Spain. Plant Disease 98: 1435. [DOI] [PubMed] [Google Scholar]
- Lombard L, Van der Merwe NA, Groenewald JZ, et al. 2015. Generic concepts in Nectriaceae. Studies in Mycology 80: 189–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Berges MS, Hera C, Sulyok M, et al. 2013. The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Molecular Microbiology 87: 49–65. [DOI] [PubMed] [Google Scholar]
- López-Díaz C, Rahjoo V, Sulyok M, et al. 2018. Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Molecular Plant Pathology 19: 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori GA, Petiet PM, Malbrán I, et al. 2012. Fusarium wilt of cyclamen: Pathogenicity and vegetative compatibility groups structure of the pathogen in Argentina. Crop Protection 36: 43–48. [Google Scholar]
- Louvet J, Toutain G. 1981. Bayoud, Fusarium wilt of date. In: Nelson PE, Toussoun TA, Cook RJ. (eds), Fusarium: Diseases, biology, and taxonomy: 13–20. The Pennsylvania State University Press, Pennsylvania State University, USA. [Google Scholar]
- Luongo L, Ferrarini A, Haegi A, et al. 2014. Genetic diversity and pathogenicity of Fusarium oxysporum f. sp. melonis races from different areas in Italy. Journal of Phytopathology 163: 73–83. [Google Scholar]
- Ma LJ, Geiser DM, Proctor RH, et al. 2013. Fusarium pathogenomics. Annual Review of Microbiology 67: 399–416. [DOI] [PubMed] [Google Scholar]
- Ma LJ, Shea T, Young S, et al. 2014. Genome sequence of Fusarium oxysporum f. sp. melonis strain NRRL 26406, a fungus causing wilt disease on melon. Genome Announcements 2: e00730–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LJ, Van der Does HC, Brokovich KA, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malençon G. 1934. Nouvelles observations concernant l’étiologie du Bayoud. Comptes rendus hebdomadaires des séances de l’Acadédes sciences, Paris 198: 1259–1261. [Google Scholar]
- Manici LM, Caputo F, Saccà ML. 2017. Secondary metabolites released into the rhizosphere by Fusarium oxysporum and Fusarium spp., as underestimated component of nonspecific replant disease. Plant and Soil 415: 85–98. [Google Scholar]
- Manicom BQ, Baayen RP. 1993. Restriction fragment length polymorphism in Fusarium oxysporum f. sp. dianthi and other fusaria from Dianthus species. Plant Pathology 42: 851–857. [Google Scholar]
- Manicom BQ, Bar-Joseph M, Kotze JM, et al. 1990. A restriction fragment length polymorphism probe relating vegetative compatibility groups and pathogenicity in Fusarium oxysporum f. sp. dianthi. Phytopathology 80: 336–339. [Google Scholar]
- Manulis S, Kogan N, Reuven M, et al. 1994. Use of the RAPD technique for identification of Fusarium oxysporum f. sp. dianthi from carnation. Phytopathology 84: 98–101. [Google Scholar]
- Marasas WFO, Nelson PE, Toussoun TA. 1984. Toxigenic Fusarium species: Identity and mycotoxicology. The Pennsylvania State University Press, University Park, Pennsylvania, USA. [Google Scholar]
- Marlatt ML, Correll JC, Kaufmann P, et al. 1996. Two genetically distinct populations of Fusarium oxysporum f. sp. lycopersici race 3 in the United States. Plant Disease 80: 1336–1342. [Google Scholar]
- Martyn RD. 1987. Fusarium oxysporum f.sp. niveum race 2: a highly aggressive race new to the United States. Plant Disease 71: 233–236. [Google Scholar]
- Martyn RD, Bruton BD. 1989. An initial survey of the United States for races of Fusarium oxysporum f.sp. niveum. HortScience 24: 696–698. [Google Scholar]
- Maryani N, Lombard L, Poerba YS, et al. 2019. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Studies in Mycology 92: 155–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marziano F, Aloj B, Noviello C. 1987. Annali della Facolta di Scienze Agrarie della Universita degli Studi di Napoli Portici 21: 13–19. [Google Scholar]
- Mason-Gamer R, Kellogg E. 1996. Testing for phylogenetic conflict among molecular datasets in the tribe Triticeae (Graminae). Systematic Biology 45: 524–545. [Google Scholar]
- Massey LM. 1926. Fusarium rot of gladiolus corms. Phytopathology 16: 509–523. [Google Scholar]
- Matuo T, Ishigami K. 1958. On the wilt of Solanum melongena L. and its causal fungus Fusarium oxysporum f. melongenae n. f. Annals of the Phytopathological Society of Japan 23: 189–192. [Google Scholar]
- Matuo T, Matsuda A, Ozaki K, et al. 1975. Fusarium oxysporum f. sp. arctii n. f. causing wilt of Great Burdock. Annuals of the Phytopathological Society of Japan 41: 77–80. [Google Scholar]
- Matuo T, Miyagawa T, Saito H. 1986. Fusarium oxysporum f. sp. garlic n. f. sp. causing basal rot of garlic. Annals of the Phytopathological Society of Japan 52: 860–864. [Google Scholar]
- Matuo T, Motohashi S. 1967. On Fusarium oxysporum f. sp. lactucae n. f. causing root rot of lettuce. Transactions of the Mycological Society of Japan 8: 13–15. [Google Scholar]
- Matuo T, Sato K. 1962. On two new forms of Fusarium lateritium. Transactions of the Mycological Society of Japan 3: 120–126. [Google Scholar]
- Matuo T, Tooyama A, Isaka M. 1979. Fusarium basal rot of Allium bakeri Regel and its causal fungus, Fusarium oxysporum Schl. f. sp. allii n.f. Annals of the Phytopathological Society of Japan 45: 305–312. [Google Scholar]
- Matuo T, Yamamoto I. 1967. On Fusarium oxysporum f. sp. lagenariae n. f. causing wilt of Lagenaria vulgaris var. hispida. Transactions of the Mycological Society of Japan 8: 61–63. [Google Scholar]
- Mbofung GY, Hong SG, Pryor BM. 2007. Phylogeny of Fusarium oxysporum f. sp. lactucae inferred from mitochondrial small subunit, elongation factor 1-α, and nuclear ribosomal intergenic spacer sequence data. Phytopathology 97: 87–98. [DOI] [PubMed] [Google Scholar]
- Mbofung GCY, Pryor BM. 2010. A PCR-based assay for detection of Fusarium oxysporum f. sp. lactucae in lettuce seed. Plant Disease 94: 860–866. [DOI] [PubMed] [Google Scholar]
- McFadden HG, Wilson IW, Chapple RM, et al. 2006. Fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) genes expressed during infection of cotton (Gossypium hirsutum). Molecular Plant Pathology 7: 87–101. [DOI] [PubMed] [Google Scholar]
- McNeill J, Turland NJ, Barrie FR, et al. (eds). 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). Gantner Verlag KG [Regnum Vegetabile no. 154]. [Google Scholar]
- McRitchie JJ. 1973. Pathogenicity and control of Fusarium oxysporum wilt of variegated Pyracantha. Plant Disease Reporter 57: 389–391. [Google Scholar]
- Mercier S, Louvet J. 1973. Recherches sur les fusarioses. X. Une fusariose vasculaire (Fusarium oxysporum) du palmier des Canaries (Phoenix canariensis). Annales de Phytopathologie 5: 203–211. [Google Scholar]
- Mes JJ, Van Doorn J, Roebroeck EJA, et al. 1994. Restriction fragment length polymorphisms, races and vegetative compatibility groups within a worldwide collection of Fusarium oxysporum f. sp. gladioli. Plant Pathology 43: 362–370. [Google Scholar]
- Mes JJ, Weststeijn EA, Herlaar F, et al. 1998. Biological and molecular characterization of Fusarium oxysporum f. sp. lycopersici divides race 1 isolates into separate virulence groups. Phytopathology 89: 156–160. [DOI] [PubMed] [Google Scholar]
- Mirocha CJ, Abbas HK, Kommedahl T, et al. 1989. Mycotoxin production by Fusarium oxysporum and Fusarium sporotrichioides isolated from Baccharis spp. from Brazil. Applied and Environmental Microbiology 55: 254–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirtalebi M, Banihashemi Z. 2014. Genetic relationship among Fusarium oxysporum f. sp. melonis vegetative compatibility groups and their relatedness to other F. oxysporum formae speciales. Journal of Agricultural Science and Technology 16: 931–943. [Google Scholar]
- Mishra RK, Pandey BK, Muthukumar M, et al. 2013a. Detection of Fusarium wilt pathogens of Psidium guajavae L. in soil using culture independent PCR (ciPCR). Saudi Journal of Biological Sciences 20: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Pandey BK, Singh V, et al. 2013b. Molecular detection and genotyping of Fusarium oxysporum f. sp. psidii isolates from different agro-ecological regions of India. Journal of Microbiology 51: 405–412. [DOI] [PubMed] [Google Scholar]
- Mishra RK, Pandey BK, Singh V, et al. 2013c. Genetic characterization of Fusarium oxysporum isolated from guava in northern India. African Journal of Microbiology Research 7: 4228–4234. [Google Scholar]
- Mishra RK, Verma DK, Pandey BK, et al. 2014. Direct colony nested-PCR for the detection of Fusarium oxysporum f. sp. psidii causing wilt disease in Psidium guajavae L. Journal of Horticulture 1: 105. doi: 10.4172/2376-0354.1000105. [DOI] [Google Scholar]
- Mohammadi N, Goltapeh EM, Babaie-Ahari A, et al. 2011. Pathogenic and genetic characterization of Iranian isolates of Fusarium oxysporum f. sp. lentis by ISSR analysis. International Journal of Agriculture Technology 7: 63–72. [Google Scholar]
- Molnár A, Sulyok L, Hornok L. 1990. Parasexual recombination between vegetative incompatible strains in Fusarium oxysporum. Mycological Research 94: 393–398. [Google Scholar]
- Moricca S, Ragazzi A, Kasuga T, et al. 1998. Detection of Fusarium oxysporum f. sp. vasinfectum in cotton tissue by polymerase chain reaction. Plant Pathology 47: 486–494. [Google Scholar]
- Mostert D, Molina AB, Daniells J, et al. 2017. The distribution and host range of the banana Fusarium wilt fungus, Fusarium oxysporum f. sp. cubense, in Asia. PLoS ONE 12 (7): e0181630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan G, Kang SW, Nam MH, et al. 2006. Characterization of Fusarium oxysporum f. sp. fragariae based on vegetative compatibility group, random amplified polymorphic DNA and pathogenicity. Plant Pathology Journal 22: 222–229. [Google Scholar]
- Nagarajan G, Nam MH, Song JY, et al. 2004. Genetic variation in Fusarium oxysporum f. sp. fragariae populations based on RAPD and rDNA RFLP analyses. Plant Pathology Journal 20: 264–270. [Google Scholar]
- Namiki F, Matsunaga M, Okuda M, et al. 2001. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Molecular Plant-Microbe Interactions 14: 580–584. [DOI] [PubMed] [Google Scholar]
- Namiki F, Shiomi T, Kayamura T, et al. 1994. Characterization of the formae speciales of Fusarium oxysporum causing wilts of cucurbits by DNA fingerprinting with nuclear repetitive DNA sequences. Applied and Environmental Microbiology 60: 2684–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki F, Shiomi T, Nishi K, et al. 1998. Pathogenic and genetic variation in the Japanese strains of Fusarium oxysporum f. sp. melonis. Phytopathology 88: 804–810. [DOI] [PubMed] [Google Scholar]
- Nash SN, Snyder WC. 1962. Quantitative estimations by plate counts of propagules of the bean rot Fusarium in field soils. Phytopathology 73: 458–462. [Google Scholar]
- Nawade B, Talaviya JR, Vyas UM, et al. 2017. Diversity analysis among Fusarium oxysporum f. sp. cumini isolates using ISSR markers, spore morphology and pathogenicity. International Journal of Current Microbiology and Applied Sciences 6: 79–87. [Google Scholar]
- Nelson PE, Toussoun TA, Marasas WFO. 1983. Fusarium species: An illustrated manual for identification. The Pennsylvania State University Press, Pennsylvania, USA. [Google Scholar]
- Netzer D. 1976. Physiological races and soil population level of Fusarium wilt of watermelon. Phytoparasitica 4: 131–136. [Google Scholar]
- Netzer D, Weintal C. 1987. Fusarium wilt of common heliotrope (Heliotropium europaeum). Phytoparasitica 15: 139–140. [Google Scholar]
- Ninet BI, Bontems JO, Lechenne O, et al. 2005. Molecular identification of Fusarium species in onychomycoses. Dermatology 210: 21–25. [DOI] [PubMed] [Google Scholar]
- Nirenberg HI. 1976. Unterschungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. [Google Scholar]
- Nirenberg HI, Ibrahim G, Michail SH. 1994. Race identity of three isolates of Fusarium oxysporum Schlecht. f. sp. vasinfectum (Atk.) Snyd. & Hans, from Egypt and the Sudan. Journal of Plant Diseases and Protection 101: 594–597. [Google Scholar]
- Nirmaladevi D, Venkataramana M, Srivastava RK, et al. 2016. Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp. lycopersici. Scientific Reports 6: 21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Kudo K. 1994. Fusarium oxysporum f. sp. colocasiae n. f. sp. causing dry rot of taro (Colocasia esculenta). Annals of the Phytopathological Society of Japan 60: 448–453. [Google Scholar]
- Nitschke E, Nihlgard M, Varrelmann M. 2009. Differentiation of eleven Fusarium spp. isolated from sugar beet, using restriction fragment analysis of a polymerase chain reaction – amplified translation elongation factor 1α gene fragment. Phytopathology 99: 921–929. [DOI] [PubMed] [Google Scholar]
- Nourollahi K, Madahjalai M. 2017. Analysis of population genetic structure of Iranian Fusarium oxysporum f. sp. lentis isolates using microsatellite markers. Australasian Plant Pathology 46: 35–42. [Google Scholar]
- Noviello C, Snyder WC. 1962. Fusarium wilt of hemp. Phytopathology 52: 1315–1317. [Google Scholar]
- Nylander JAA. 2004. MrModeltest v. 2. Programme distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Cigelnik E. 1999. A DNA sequence-based phylogenetic structure for the Fusarium oxysporum complex. Phytoparasitica 27: 69. [Google Scholar]
- O’Donnel K, Gueidan C, Sink S, et al. 2009. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genetics and Biology 46: 936–948. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, et al. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Rooney AP, Proctor RH, et al. 2013. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genetics and Biology 52: 20–31. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Sarver BAJ, Brandt M, et al. 2007. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreak of 2005 and 2006. Journal of Clinical Microbiology 45: 2235–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Fothergill A, et al. 2008. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. Journal of Clinical Microbiology 46: 2477–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, et al. 2004. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: Evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. Journal of Clinical Microbiology 42: 5109–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, et al. 2010. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. Journal of Clinical Microbiology 48: 3708–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Ward TJ, Robert VARG, et al. 2015. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica 43: 583–595. [Google Scholar]
- Ochoa JB, Yangari BF, Ellis MA, et al. 2004. Two new formae specialis of Fusarium oxysporum, causing vascular wilt on babaco (Carica heilbornii var. pentagona) and vascular wilt on naranjilla (Solanum quitoense) in Ecuador. Fitologiya 39: 10–17. [Google Scholar]
- Ogiso H, Fujinaga M, Saito H, et al. 2002. Physiological races and vegetative compatibility groups of Fusarium oxysporum f. sp. lactucae isolated from crisphead lettuce in Japan. Journal of General Plant Pathology 68: 292–299. [Google Scholar]
- Okubara PA, Harrison LA, Gatch EW, et al. 2013. Development and evaluation of a TaqMan real-time PCR assay for Fusarium oxysporum f. sp. spinaciae. Plant Disease 97: 927–937. [DOI] [PubMed] [Google Scholar]
- Okuda M, Ikeda K, Namiki F, et al. 1998. Tfo1: an Ac-like transposon from the plant pathogenic fungus Fusarium oxysporum. Molecular Genetics and Genomics 258: 599–607. [DOI] [PubMed] [Google Scholar]
- Ortiz CS, Bell AA, Magill CW, et al. 2017. Specific PCR detection of Fusarium oxysporum f. sp. vasinfectum California race 4 based on a unique Tfo1 insertion event in the PHO gene. Plant Disease 101: 34–44. [DOI] [PubMed] [Google Scholar]
- Ortu G, Bertetti D, Gullino ML, et al. 2013. A new forma specialis of Fusarium oxysporum on Crassula ovata. Journal of Plant Pathology 95: 33–39. [Google Scholar]
- Ortu G, Bertetti D, Gullino ML, et al. 2015a. Fusarium oxysporum f. sp. echeveriae, a novel forma specialis causing crown and stem rot of Echeveria agavoides. Phytopathologia Mediterranea 54: 64–75. [Google Scholar]
- Ortu G, Bertetti D, Martin P, et al. 2015b. Fusarium oxysporum f. sp. papaveris: a new formae specialis isolated from Iceland poppy (Papaver nudicaule). Phytopathologia Mediterranea 54: 76–85. [Google Scholar]
- Owen JH. 1956. Cucumber wilt, caused by Fusarium oxysporum f. cucumerinum n. f. Phytopathology 46: 153–157. [Google Scholar]
- Padwick GW. 1940. The genus Fusarium. III. A critical study of the fungus causing wilt of gram (Cicer arietinum L.) and of the related species in the sub-section Orthocera, with special relation to variability of key characters. Indian Journal of Agricultural Sciences 10: 241–284. [Google Scholar]
- Palmero D, Rubio-Moraga A, Galvez-Patón L, et al. 2014. Pathogenicity and genetic diversity of Fusarium oxysporum isolates from corms of Crocus sativus. Industrial Crops and Products 61: 186–192. [Google Scholar]
- Pande A, Rao VG. 1990. Wilt disease of Tabernaemontana coronaria Willd. Biovigyanam 16: 58–61. [Google Scholar]
- Pandotra VR, Gupta JH, Sastry KSM. 1971. Note on wilt disease of Solanum laciniatum. Indian Journal of Mycology and Plant Pathology 1: 86–87. [Google Scholar]
- Pappalardo L, Smith MK, Hamill SD, et al. 2009. DNA amplification fingerprinting analysis of genetic variation within Fusarium oxysporum f. sp. zingiberi. Australasian Plant Pathology 38: 51–54. [Google Scholar]
- Pasquali M, Acquadro A, Balmas V, et al. 2003. RAPD characterization of Fusarium oxysporum isolates pathogenic on Argyranthemum frutescens L. Journal of Phytopathology 151: 30–35. [Google Scholar]
- Pasquali M, Acquadro A, Balmas V, et al. 2004a. Development of PCR primers for a new Fusarium oxysporum pathogenic on Paris daisy (Argyranthemum frutescens L.). European Journal of Plant Pathology 110: 7–11. [Google Scholar]
- Pasquali M, Dematheis F, Gilardi G, et al. 2005. Vegetative compatibility groups of Fusarium oxysporum f. sp. lactucae from lettuce. Plant Disease 89: 237–240. [DOI] [PubMed] [Google Scholar]
- Pasquali M, Dematheis F, Gullino ML, et al. 2007. Identification of race 1 of Fusarium oxysporum f. sp. lactucae on lettuce by inter-retrotransposon sequence-characterized amplified region technique. Phytopathology 97: 987–996. [DOI] [PubMed] [Google Scholar]
- Pasquali M, Marena L, Fiora E, et al. 2004b. Real-time polymerase chain reaction for identification of highly pathogenic group of Fusarium oxysporum f. sp. chrysanthemi on Argyranthemum frutescens L. Journal of Plant Pathology 86: 53–59. [Google Scholar]
- Pasquali M, Marena L, Gullino ML, et al. 2004c. Vegetative compatibility grouping of the Fusarium wilt pathogen of Paris daisy (Argyranthemum frutescens L.). Journal of Phytopathology 152: 257–259. [Google Scholar]
- Pasquali M, Piatti P, Gullino ML, et al. 2006. Development of a real-time polymerase chain reaction for the detection of Fusarium oxysporum f. sp. basilici from basil seeds and roots. Journal of Phytopathology 154: 632–636. [Google Scholar]
- Pasquali M, Saravanakumar D, Gullino ML, et al. 2008. Sequence-specific amplified polymorphism (SSAP) technique to analyse Fusarium oxysporum f. sp. lactucae VCG 0300 isolate from lettuce. Journal of Plant Pathology 90: 527–535. [Google Scholar]
- Pastrana AM, Kirkpatrick SC, Kong M, et al. 2017. Fusarium oxysporum f. sp. mori, a new forma specialis causing Fusarium wilt of blackberry. Plant Disease 100: 1018. [DOI] [PubMed] [Google Scholar]
- Patel PN, Prasad N, Mathur RL, et al. 1957. Fusarium wilt of cumin. Current Science 26: 181–182. [Google Scholar]
- Pinaria AG, Laurence MH, Burgess LW, et al. 2015. Phylogeny and origin of Fusarium oxysporum f. sp. vanillae in Indonesia. Plant Pathology 64: 1358–1365. [Google Scholar]
- Pintore I, Gilardi G, Gullino ML, et al. 2017. Analysis of vegetative compatibility groups of Italian and Dutch isolates of Fusarium oxysporum f. sp. lactucae. Journal of Plant Pathology 99: 517–521. [Google Scholar]
- Ploetz RC. 2006. Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp. cubense. Phytopathology 96: 653–656. [DOI] [PubMed] [Google Scholar]
- Ploetz RC. 2015. Fusarium wilt of banana. Phytopathology 105: 1512–1521. [DOI] [PubMed] [Google Scholar]
- Poli A, Bertetti D, Rapetti S, et al. 2013. Characterization and identification of Colombian isolates of Fusarium oxysporum f. sp. dianthi. Journal of Plant Pathology 95: 255–263. [Google Scholar]
- Poli A, Gilardi G, Spadaro D, et al. 2012. Molecular characterization of Fusarium oxysporum f. sp. cichorii pathogenic on chicory (Cichorium intybus). Phytoparasitica 40: 383–391. [Google Scholar]
- Pouralibaba HR, Pérez-de-Luque A, Rubiales D. 2017. Histopathology of the infection on resistance and susceptible lentil accessions by two contrasting pathotypes of Fusarium oxysporum f. sp. lentis. European Journal of Plant Pathology 148: 53–63. [Google Scholar]
- Pouralibaba HR, Rubiales D, Fondevilla S. 2016. Identification of pathotypes in Fusarium oxysporum f. sp. lentis. European Journal of Plant Pathology 144: 539–549. [Google Scholar]
- Prasad MSL, Sujatha M, Raoof MA. 2008. Morphological, pathogenic and genetic variability in castor wilt isolates. Indian Phytopathology 61: 18–27. [Google Scholar]
- Prasad N, Mehta PR, Lal SB. 1952. Fusarium wilt of guava (Psidium guajava L.) in Uttar Pradesh, India. Nature 169: 753–754. [DOI] [PubMed] [Google Scholar]
- Puhalla JE. 1984a. A visual indicator of heterokaryosis in Fusarium oxysporum from celery. Canadian Journal of Botany 62: 540–545. [Google Scholar]
- Puhalla JE. 1984b. Races of Fusarium oxysporum f. sp. apii in California and their genetic interrelationships. Canadian Journal of Botany 62: 546–550. [Google Scholar]
- Puhalla JE. 1985. Classification of strains of Fusarium oxysporum on the basis of vegetative compatibility. Canadian Journal of Botany 63: 179–183. [Google Scholar]
- Pyler TR, Simone GW, Fernandez D, et al. 2000. Genetic diversity among isolates of Fusarium oxysporum f. sp. canariensis. Plant Pathology 49: 155–164. [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ, et al. 2014. Introducing the Consolidated Species Concepts to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe RD. 1960. Fusarium wilt of Sedum. Phytopathology 50: 651. [Google Scholar]
- Raabe RD. 1985a. Fusarium wilt of Eustoma grandiflora. Phytopathology 75: 1306. [Google Scholar]
- Raabe RD. 1985b. Fusarium wilt of Hebe species. Plant Disease 69: 450–451. [Google Scholar]
- Rafique K, Rauf CA, Naz F, et al. 2015. DNA sequence analysis, morphology and pathogenicity of Fusarium oxysporum f. sp. lentis isolates inciting lentil wilt in Pakistan. International Journal of Biosciences 7: 74–91. [Google Scholar]
- Ramirez-Villupadua J, Endo RM, Bosland P, et al. 1985. A new race of Fusarium oxysporum f. sp. conglutinans that attacks cabbage with type A resistance. Plant Disease 69: 612–613. [Google Scholar]
- Rataj-Guranowska M, Wiatroszak I, Hornok L. 1984. Serological comparison of two races of Fusarium oxysporum f. sp. lupini. Journal of Phytopathology 110: 221–225. [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. CMI and British Mycological Society, Kew, Surrey, UK. [Google Scholar]
- Reddy JM, Raoof MA, Ulaganathan K. 2012. Development of specific markers for identification of Indian isolates of Fusarium oxysporum f. sp. ricini. European Journal of Plant Pathology 134: 713–719. [Google Scholar]
- Reid J. 1958. Studies on the fusaria which cause wilt in melons. 1. The occurrence and distribution of races of the muskmelon and watermelon fusaria and a histological study of the colonization of muskmelon plants susceptible or resistant to Fusarium wilt. Canadian Journal of Botany 36: 393–410. [Google Scholar]
- Ren Y, Jiao D, Gong G, et al. 2015. Genetic analysis and chromosome mapping of resistance to Fusarium oxysporum f. sp. niveum (FON) race 1 and race 2 in watermelon (Citrullus lanatus L.). Molecular Breeding 35: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheeder JP, Marasas WFO, Vismer HF. 2002. Production of fumonisins analogs by Fusarium species. Applied and Environmental Microbiology 68: 2101–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro RLD. 1977. Race differentiation in Fusarium oxysporum f. sp. phaseoli, the causal agent of bean yellows. Proceeding of the American Phytopathology Society 4: 165. [Google Scholar]
- Ribeiro RLD, Hagedorn DJ. 1979. Screening for resistance to and pathogenic specialization of Fusarium oxysporum f. sp. phaseoli, the causal agent of bean yellows. Phytopathology 69: 272–276. [Google Scholar]
- Richter H. 1941. Lupinenfusariosen. Mitteilungen aus der Biologischen Reichanstalt fur Land-und Forstwirtschaft 64: 50–61. [Google Scholar]
- Risser G, Banihashemi Z, Davis DW. 1976. A proposed nomenclature of Fusarium oxysporum f. sp. melonis races and resistance genes in Cucumis melo. Phytopathology 66: 1105–1106. [Google Scholar]
- Risser G, Mas P. 1965. Mise en évidencede plusieurs races de Fusarium oxysporum f. melonis. Annales de l’amélioration des plantes 15: 405–408. [Google Scholar]
- Roebroeck EJA. 2000. Fusarium oxysporum from iridaceous crops: analysis of genetic diversity and host specialisation. Faculty of Science, University of Amsterdam, The Netherlands. [Google Scholar]
- Roebroeck EJA, Mes JJ. 1992. Physiological races and vegetative compatibility groups within Fusarium oxysporum f. sp. gladioli. Netherlands Journal of Plant Pathology 98: 57–64. [Google Scholar]
- Romano C, Miracco C, Difonzo EM. 1998. Skin and nail infections due to Fusarium oxysporum in Tuskany, Italy. Mycoses 41: 433–437. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rosewich UL, Pettway RE, Katan T, et al. 1999. Population genetic analysis corroborates dispersal of Fusarium oxysporum f. sp. radicis-lycopersici from Florida to Europe. Phytopathology 89: 623–630. [DOI] [PubMed] [Google Scholar]
- Salgado MO, Schwartz HF. 1993. Physiological specialization and effects of inoculum concentration of Fusarium oxysporum f. sp. phaseoli on common beans. Plant Disease 77: 492–496. [Google Scholar]
- Sandoval-Denis M, Guarnaccia V, Polizzi G, et al. 2018. Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 40: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands DC, Ford EJ, Miller RV, et al. 1997. Characterization of a vascular wilt of Erythroxylum coca caused by Fusarium oxysporum f. sp. erythroxyli forma specialis nova. Plant Disease 81: 501–504. [DOI] [PubMed] [Google Scholar]
- Sauthoff W, Gerlach W. 1957. Die Fusarium – Welke der Aechmea fasciata – eine neue pilzkrankheit. Gartenwelt 23: 389–390. [Google Scholar]
- Sauthoff W, Gerlach W. 1958. Über eine bisher nicht bekannte Fusarium – welkekrankheit an Aechmea fasciata (Lindl). Bak. Nachrichtenblatt des Deutschen Pflanzenschutzdienstes (Braunschweig) 10: 1–3. [Google Scholar]
- Scarlett K, Tesoriero L, Daniel R, et al. 2013. Detection and quantification of Fusarium oxysporum f. sp. cucumerinum in environmental samples using specific quantitative PCR assay. European Journal of Plant Pathology 137: 315–324. [Google Scholar]
- Schmidt SM, Lukasiewicz J, Farrer R, et al. 2016. Comparative genomics of Fusarium oxysporum f. sp. melonis reveals the secreted protein recognized by the Fom-2 resistance gene in melon. New Phytologist 209: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider RW, Norelli JL. 1981. A new race of Fusarium oxysporum f. sp. apii in California. Phytopathology 71: 108. [Google Scholar]
- Schreuder JC. 1951. Een onderzoek over de Amerikaanse vaatziekte van de erwten in Nederland. Tijdschrift over Plantenziekten 57 (6): 175–206. [Google Scholar]
- Sebastiani MS, Bagnaresi P, Sestili S, et al. 2017. Transcriptome analysis of the melon- Fusarium oxysporum f. sp. melonis race 1.2 pathosystem in susceptible and resistant plants. Frontiers in Plant Science 8: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergent E, Beguet M. 1921. Sur la nature mycosique d’une nouvelle maladies des dattiers menaçant les oasis marocaines. Comptes rendus hebdomadaires des séances de l’Acadédes sciences, Paris 172: 1624–1627. [Google Scholar]
- Sharma KD, Winter P, Kahl G, et al. 2004. Molecular mapping of Fusarium oxysporum f. sp. ciceris race 3 resistance gene in chickpea. Theoretical and Applied Genetics 108: 1243–1248. [DOI] [PubMed] [Google Scholar]
- Sharma M, Nagavardhini A, Thudi M, et al. 2014. Development of DArT markers and assessment of diversity in Fusarium oxysporum f. sp. ciceris, wilt pathogen of chickpea (Cicer arietinum L.). BMC Genomics 15: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Sengupta A, Ghosh R, et al. 2016. Genome wide transcriptome profiling of Fusarium oxysporum f. sp. ciceris conidial germination reveals new insights into infection-related genes. Science Reports 6: 37353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shende SS, Wankhade DJ, Rajurkar AB, et al. 2015. Characterization of pathogenic species of Fusarium oxysporum isolates of safflower by RAPD marker. Indian Journal of Plant Protection 43: 251–253. [Google Scholar]
- Shimazu J, Yamauchi N, Hibi T, et al. 2005. Development of sequence tagged site markers to identify races of Fusarium oxysporum f. sp. lactucae. Journal of General Plant Pathology 71: 183–189. [Google Scholar]
- Shiraishi A, Leslie JF, Zhong S, et al. 2012. ALFP, pathogenicity, and VCG analyses of Fusarium oxysporum and Fusarium pseudocircinatum from Acacia koa. Plant Disease 96: 1111–1117. [DOI] [PubMed] [Google Scholar]
- Skovgaard K, Nirenberg HI, O’Donnell K, et al. 2001. Evolution of Fusarium oxysporum f. sp. vasinfectum races inferred from multigene genealogies. Phytopathology 91: 1231–1237. [DOI] [PubMed] [Google Scholar]
- Smith EF. 1910. A Cuban banana disease. Science, New Series 31: 754–755. [Google Scholar]
- Smith SN, DeVay JE, Hsieh WH, et al. 2001. Soil-borne populations of Fusarium oxysporum f. sp. vasinfectum, a cotton wilt fungus in California fields. Mycologia 93: 737–743. [Google Scholar]
- Smith SN, Helms DM, Temple SR, et al. 1999. The distribution of Fusarium wilt of blackeyed cowpeas within California caused by Fusarium oxysporum f. sp. tracheiphilum race 4. Plant Disease 83: 694. [DOI] [PubMed] [Google Scholar]
- Snyder WC, Hansen HN. 1940. The species concept in Fusarium. American Journal of Botany 27: 64–67. [Google Scholar]
- Snyder WC, Toole ER, Hepting GH. 1949. Fusaria associated with mimosa wilt, and pine pitch canker. Journal of Agricultural Research 78: 365–382. [Google Scholar]
- Snyder WC, Walker JC. 1935. Fusarium near wilt of pea. Zentralblatt für Bakteriologie, Parasitenkunde und Infektionskrankheiten Abteilung II, Jena 91: 355–378. [Google Scholar]
- Southwood MJ, Viljoen A, Mostert G, et al. 2012. Molecular identification of two vegetative compatibility groups of Fusarium oxysporum f. sp. cepae. Phytopathology 102: 204–213. [DOI] [PubMed] [Google Scholar]
- Srinivasan K, Gilardi G, Spadaro D, et al. 2010. Molecular characterization through IGS sequencing of formae speciales of Fusarium oxysporum pathogenic on lamb’s lettuce. Phytopathologia Mediterrania 49: 309–320. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg C, Laurent J, Edel-Hermann V, et al. 2015. Adaptation of Fusarium oxysporum and Fusarium dimerum to the specific aquatic environment provided by the water systems of hospitals. Water Research 76: 53–65. [DOI] [PubMed] [Google Scholar]
- Stewart D. 1931. Sugar beet yellows caused by Fusarium conglutinane var. betae. Phytopathology 19: 59–70. [Google Scholar]
- Stover RH. 1962. Fusarial Wilt (Panama Disease) of bananas and other Musa species. Commonwealth Mycological Institute, Kew, England. [Google Scholar]
- Suelong H. 1981. A study on the pathogen of blight disease of tungoil trees. Journal of Nanjing Technological College of Forest Products 9: 53. [Google Scholar]
- Suga H, Hirayama Y, Morishima M, et al. 2013. Development of PCR primers to identify Fusarium oxysporum f. sp. fragariae. Plant Disease 97: 619–625. [DOI] [PubMed] [Google Scholar]
- Summerell BA, Leslie JF, Liew ECY, et al. 2010. Fusarium species associated with plants in Australia. Fungal Diversity 46: 1–27. [Google Scholar]
- Sun SK, Huang JW. 1983. A new Fusarium wilt of bitter gourd in Taiwan. Plant Disease 67: 226–227. [Google Scholar]
- Sung GH, Sung JM, Hywel-Jones NL, et al. 2007. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Molecular phylogenetics and evolution 44: 1204–1223. [DOI] [PubMed] [Google Scholar]
- Swanson TA, Van Gundy SD. 1985. Influences of temperature and plant age on differentiation of races of Fusarium oxysporum f. sp. tracheiphilum on cowpea. Plant Disease 69: 779–781. [Google Scholar]
- Swett CS, Uchida JY. 2015. Characterization of Fusarium diseases on commercially grown orchids in Hawaii. Plant Pathology 64: 648–654. [Google Scholar]
- Swift CE, Wickliffe ER, Schwartz HF. 2002. Vegetative compatibility groups of Fusarium oxysporum f. sp. cepae from onion in Colorado. Plant Disease 86: 606–610. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), v. 4.0b10. Computer programme. Sinauer Associates, Sunderland, Massachusetts, USA. [Google Scholar]
- Taheri N, Rastegar MF, Jafarpour B, et al. 2010. Pathogenic and genetic characterization of Fusarium oxysporum f. sp. lentis by RAPD and IGS analysis in Khorasan Province. World Applied Science Journal 9: 239–244. [Google Scholar]
- Takehara T, Kuniyasu K, Mori M, et al. 2003. Use of a nitrate-nonutilizing mutant and selective media to examine population dynamics of Fusarium oxysporum f. sp. spinaciae in soil. Phytopathology 93: 1173–1181. [DOI] [PubMed] [Google Scholar]
- Takken F, Rep M. 2010. The arms race between tomato and Fusarium oxysporum. Molecular Plant Pathology 11: 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaviya JR, Jadeja KB, Nawade BD. 2014. Variability among different isolates of Fusarium oxysporum f. sp. cumini. Trends in Biosciences 7: 3611–3616. [Google Scholar]
- Tantaoui A, Boisson C. 1991. Compatibilité végétative d’isolats du Fusarium oxysporum f. sp. albedinis et des Fusarium oxysporum de la rhizosphère du palmier dattier et des sols de palmeraies. Phytopathologia Mediterriana 30: 155–163. [Google Scholar]
- Tantaoui A, Fernandez D. 1993. Comparaison entre Fusarium oxysporum f. sp. albedinis et Fusarium oxysporum des sols palmeraies par l’étude du polymorphisme de longueur des fragments de restriction (RFLP). Phytopathologia Mediterriana 32: 235–244. [Google Scholar]
- Tantaoui A, Ouinten M, Geiger J-P-, et al. 1996. Characterization of a single clonal lineage of Fusarium oxysporum f. sp. albedinis causing bayoud disease of date palm in Morocco. Phytopathology 86: 787–792. [Google Scholar]
- Taylor A, Vágány V, Jackson AC, et al. 2016. Identification of pathogenicity-related genes in Fusarium oxysporum f. sp. cepae. Molecular Plant Pathology 17: 1032–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher LF, Gardiner DM, Kazan K, et al. 2012. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Molecular Plant-Microbe Interactions 25: 180–190. [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Williams AH, Grag G, et al. 2016. Transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. medicaginis during colonisation of resistant and susceptible Medicago truncatula hosts identifies differential pathogenicity profiles and novel candidate effectors. BMC Genomics 17: 860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurland NJ, Wiersma JH, Barbie FR, et al. (eds). 2018. International Code of Nomenclature for algea, fungi, and plants (Shenzhen Code). Koeltz Botanical Books. [Regnum Vegetabile 159]. [Google Scholar]
- Timmer LW. 1982. Host range and host colonization, temperature effects, and dispersal of Fusarium oxysporum f. sp. citri. Phytopathology 72: 698–702. [Google Scholar]
- Timmer LW, Garnsey SM, Grimm GR, et al. 1979. Wilt and dieback of Mexican lime caused by Fusarium oxysporum. Phytopathology 69: 730–734. [Google Scholar]
- Tok F, Kurt S. 2010. Pathogenicity, vegetative compatibility and amplified fragment length polymorphism (AFLP) analysis of Fusarium oxysporum f. radicis-cucumerinum isolates from Turkish greenhouses. Phytoparasitica 38: 253–260. [Google Scholar]
- Toole ER. 1941. Fusarium wilt of the mimosa tree (Albizzia julibrissin). Phytopathology 31: 599–616. [Google Scholar]
- Toole ER. 1952. Two races of Fusarium oxysporum f. perniciosum causing wilt of Albizzia spp. Phytopathology 42: 694. [Google Scholar]
- Toth KF, Lacy ML. 1991. Comparing vegetative compatibility and protein banding patterns for identification of Fusarium oxysporum f.sp. apii race 2. Canadian Journal Microbiology 37: 669–674. [Google Scholar]
- Triolo E, Lorenzini G. 1983. Fusarium oxysporum f. sp. fatshederae, a new forma specialis causing wilt of × Fatshedera lizei. Annals of Applied Biology 102: 245–250. [Google Scholar]
- Troisi M, Bertetti D, Gullino ML, et al. 2013. Race differentiation in Fusarium oxysporum f. sp. chrysanthemi. Journal of Phytopathology 161: 675–688. [Google Scholar]
- Troisi M, Gullino ML, Garibaldi A. 2010. Gerbera jamesonii, a new host of Fusarium oxysporum f. sp. tracheiphilum. Journal of Phytopathology 158: 8–14. [Google Scholar]
- Trujillo EE. 1963. Fusarium yellows and rhizome rot of common ginger. Phytopathology 53: 1370–1371. [Google Scholar]
- Tucker CM. 1927. Vanilla root rot. Journal of Agricultural Research 35: 1121–1136. [Google Scholar]
- Upasani ML, Gurjar GS, Kadoo NY, et al. 2016. Dynamics of colonization and expression of pathogenicity related genes in Fusarium oxysporum f. sp. ciceri during chickpea vascular wilt disease progression. PLoS ONE 11: e0156490. doi: 10.1371/journal.pone.0156490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakalounakis DJ. 1996. Root and stem rot of cucumber caused by Fusarium oxysporum f. sp. radicis-cucumerinum f. sp. nov. Plant Disease 80: 313–316. [DOI] [PubMed] [Google Scholar]
- Vakalounakis DJ, Doulis AG, Klironomou E. 2005. Characterization of Fusarium oxysporum f. sp. radicis-cucumerinum attacking melon under natural conditions in Greece. Plant Pathology 54: 339–346. [Google Scholar]
- Vakalounakis DJ, Fragkiadakis GA. 1999. Genetic diversity of Fusarium oxysporum isolates from cucumber: differentiation by pathogenicity, vegetative compatibility, and RAPD fingerprinting. Phytopathology 89: 161–168. [DOI] [PubMed] [Google Scholar]
- Vakalounakis DJ, Wang Z, Fragkiadakis GA, et al. 2004. Characterization of Fusarium oxysporum isolates obtained from cucumber in China by pathogenicity, VCG, and RAPD. Plant Disease 88: 645–649. [DOI] [PubMed] [Google Scholar]
- Van Dam P, Fokkens L, Schmidt SM, et al. 2016. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environmental Microbiology 18: 4087–4102. [DOI] [PubMed] [Google Scholar]
- Van Dam P, Rep M. 2017. The distribution of Miniature Impala elements and SIX genes in the Fusarium genus is suggestive of horizontal gene transfer. Journal of Molecular Evolution 85: 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does HC, Lievens B, Claes L, et al. 2008. The presence of the virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environmental Microbiology 10: 1475–1485. [DOI] [PubMed] [Google Scholar]
- Van Diepeningen AD, Feng P, Ahmed S, et al. 2015. Spectrum of Fusarium infections in tropical dermatology evidenced by multilocus sequencing typing diagnostics. Mycoses 58: 48–57. [DOI] [PubMed] [Google Scholar]
- Van Hall CJJ. 1903. Die Sankt Johaniskrankheit der Erbsen verursacht von Fusarium vasinfectum Atk. Berichte der Deutschen Botanischen Gesellschaft 21: 2–6. [Google Scholar]
- Vasudeva RS, Srinivasan KV. 1952. Studies on the wilt diseases of lentil. Indian Phytopathology 5: 23–32. [Google Scholar]
- Venter SL, Theron DJ, Steyn PJ, et al. 1992. Relationship between vegetative compatibility and pathogenicity of isolates of Fusarium oxysporum f. sp. tuberosi from potato. Phytopathology 82: 858–862. [Google Scholar]
- Von Arx JA. 1952. De voetziekte van Gerbera, veroorzaakt door Fusarium oxysporum Schlecht. Tijdschrift over Plantziekten 58 (1): 5–9. [Google Scholar]
- Von Schlechtendahl FK. 1824. Flora berolinensis. Pars secunda. Cryptogamia, Berlin. [Google Scholar]
- Wager VA. 1947. Wilt disease of New Zealand flax. Farming in South Africa 22: 871–878. [Google Scholar]
- Waite BH. 1963. Wilt of Heliconia spp. caused by Fusarium oxysporum f. cubense race 3. Tropical Agriculture 40: 299–305. [Google Scholar]
- Wang B, Brubaker CL, Summerell BA, et al. 2010. Local origin of two vegetative compatibility groups of Fusarium oxysporum f. sp. vasinfectum in Australia. Evolutionary Applications 3 (5–6): 505–524. doi: 10.1111/j.1752-4571.2010.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Brubaker CL, Tate W, et al. 2006. Genetic variation and population structure of Fusarium oxysporum f. sp. vasinfectum in Australia. Plant Pathology 55: 746–755. [Google Scholar]
- Wang PH, Lo HS, Yeh Y. 2001. Identification of F. o. cucumerinum and F. o. luffae by RAPD-generated DNA probes. Letters in Applied Microbiology 33: 397–401. [DOI] [PubMed] [Google Scholar]
- Webb KM, Case AJ, Brick MA, et al. 2013. Cross pathogenicity and vegetative compatibility of Fusarium oxysporum isolated from sugar beet. Plant Disease 97: 1200–1206. [DOI] [PubMed] [Google Scholar]
- Weimer JL. 1928. A wilt disease of alfalfa caused by Fusarium oxysporum var. medicaginis, n. var. Journal of Agricultural Research 37: 419–433. [Google Scholar]
- Weimer JL. 1944. Some root rots and a foot rot of lupines in the southeastern part of the United States. Journal of Agricultural Research 68: 441–457. [Google Scholar]
- Wellman FL. 1972. Tropical American plant disease (Neotropical phytopathology problems). Scarecrow Press, Metuchen, N.J., USA. [Google Scholar]
- Wellman M. 1954. Fusarium oxysporum Shl. f. sp. coffea. Phytopathology 44: 509. [Google Scholar]
- Whitehead DS, Coddington A, Lewis BG. 1992. Classification of races by DNA polymorphism analysis and vegetative compatibility grouping in Fusarium oxysporum f. sp. pisi. Physiological and Molecular Plant Pathology 41: 295–305. [Google Scholar]
- Widodo, Kondo N, Kobayashi K, et al. 2008. Vegetative compatibility groups within Fusarium oxysporum f. sp. cepae in Hokkaido-Japan. Microbiology Indonesia 2: 39–43. [Google Scholar]
- Williams AH, Sharma M, Thatcher LF, et al. 2016. Comparative genomics and prediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genomics 17: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winks BL, Williams YN. 1965. A wilt of strawberry caused by a new form of Fusarium oxysporum. Queensland Journal of Agriculture and Animal Science 22: 475–479. [Google Scholar]
- Wollenweber HW. 1913. Studies on the Fusarium problem. Phytopathology 3: 24–50. [Google Scholar]
- Wollenweber HW. 1914. Identification of species of Fusarium occurring on the sweet potato, Ipomoea batatas. Journal of Agricultural Research 2: 251–285. [Google Scholar]
- Wollenweber HW. 1931. Fusarium-monographie. Fungi parasitici et saprophytici. Zeitschrift für Parasitenkunde 3: 260–516. [Google Scholar]
- Wollenweber HW, Reinking OA. 1935. Die Fusarien, ihre Beschreibung, Schadwirkung und Bekampfung. Paul Parey, Berlin. [Google Scholar]
- Woo SL, Zoina A, Del Sorbo G, et al. 1996. Characterization of Fusarium oxysporum f. sp. phaseoli by pathogenic races, VCGs, RFLPs, and RAPD. Phytopathology 86: 966–973. [Google Scholar]
- Woudenberg JHC, Seidl MF, Groenewald JZ, et al. 2015. Alternaria section Alternata: Species, formae speciales or pathotypes? Studies in Mycology 82: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt LP, Neuvel A, Sikkema A, et al. 1995. Genetic variation in Fusarium oxysporum from cyclamen. Phytopathology 85: 1348–1355. [Google Scholar]
- Wunsch MJ, Baker AH, Kalb DW, et al. 2009. Characterization of Fusarium oxysporum f. sp. loti forma specialis nov., a monophyletic pathogen causing vascular wilt of birdsfoot trefoil. Plant Disease 93: 58–66. [DOI] [PubMed] [Google Scholar]
- Yamauchi N, Horiuchi S, Satou M. 2001. Pathogenicity groups in Fusarium oxysporum f. sp. lactucae on horticultural types of lettuce cultivars. Journal of General Plant Pathology 67: 288–290. [Google Scholar]
- Yamauchi N, Shimazu J, Satou M, et al. 2004. Physiological races and vegetative compatibility groups of butterhead lettuce isolates of Fusarium oxysporum f. sp. lactucae in Japan. Journal of General Plant Pathology 70: 308–313. [Google Scholar]
- Yoo SJ, Watanabe H, Kobayashi K, et al. 1993. Vegetative compatibility grouping of formae speciales of Fusarium oxysporum pathogenic to the Liliaceae. Annals of the Phytopathological Society of Japan 59: 3–9. [Google Scholar]
- Yu TF, Fang CT. 1948. Fusarium diseases of broad beans. III. Root-rot and wilt of broad beans caused by two new forms of Fusarium. Phytopathology 38: 587–594. [Google Scholar]
- Yun SH, Arie T, Kaneko I, et al. 2000. Molecular organization of mating type loci in heterothallic, homothalic, and asexual Gibberella/Fusarium species. Fungal Genetics and Biology 31: 7–20. [DOI] [PubMed] [Google Scholar]
- Zambounis AG, Paplomatas E, Tsaftaris AS. 2007. Intergenic spacer–RFLP analysis and direct quantification of Australian Fusarium oxysporum f. sp. vasinfectum isolates from soil and infected cotton tissues. Plant Disease 91: 1564–1573. [DOI] [PubMed] [Google Scholar]
- Zang W, Zhao W, Huang J, et al. 2014. PEG-mediated genetic transformation of Fusarium oxysporum f. sp. conglutinans to study pathogenesis in cabbage. Chiang Mai Journal Science 41: 945–956. [Google Scholar]
- Zanotti MGS, De Queiroz MV, Dos Santos JK, et al. 2006. Analysis of the genetic diversity of Fusarium oxysporum f. sp. phaseoli isolates, pathogenic and non-pathogenic to common bean (Phaseolus vulgaris L.). Journal of Phytopathology 154: 545–549. [Google Scholar]
- Zhang Z, Zhang J, Wang Y, et al. 2005. Molecular detection of Fusarium oxysporum f. sp. niveum and Mycosphaerella melonis in infected plant tissues and soil. FEMS Microbiology Letters 249: 39–47. [DOI] [PubMed] [Google Scholar]
- Zhou XG, Everts KL, Bruton BD. 2010. Race 3, a new and highly virulent race of Fusarium oxysporum f. sp. niveum causing Fusarium wilt in watermelon. Plant Disease 94: 92–98. [DOI] [PubMed] [Google Scholar]
- Zimmermann J, De Klerk M, Musyoki MK, et al. 2015. An explicit AFLP-based marker for monitoring Fusarium oxysporum f. sp. strigae in tropical soils. Biological Control 89: 42–52. [Google Scholar]
- Zimmermann J, Musyoki MK, Cadisch G, et al. 2016. Proliferation of the biocontrol agent Fusarium oxysporum f. sp. strigae and its impact on indigenous rhizosphere fungal communities maize under different agro-ecologies. Rhizosphere 1: 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]