Fig. 1.
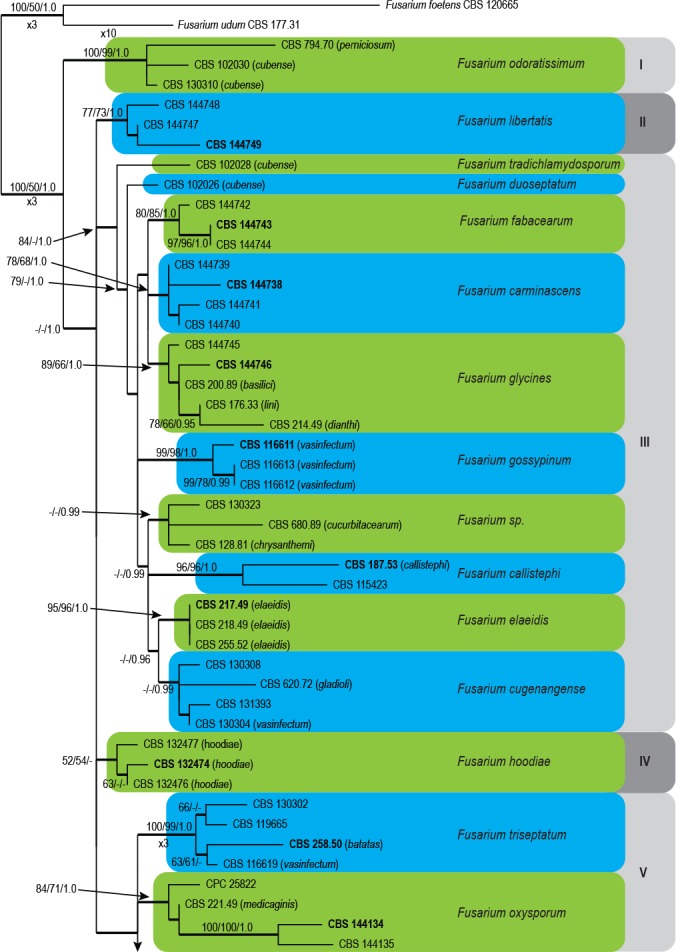
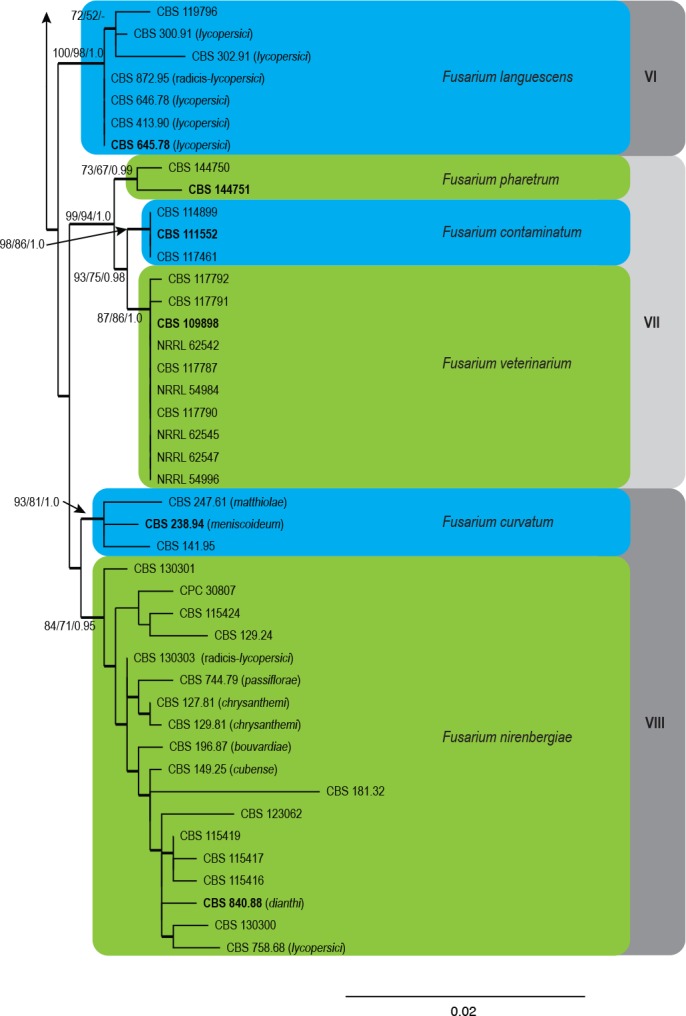
The ML consensus tree inferred from the combined cmdA, rpb2, tef1 and tub2 sequence alignment. Thickened branches indicate branches present in the ML, MP and Bayesian consensus trees. Support values (ML & MP bootstrap and posterior probability values) are indicated at the branches. The scale bar indicates 0.02 expected changes per site. Clade numbers are provided on the right of the tree and these are used for reference in the treatment of the species. The tree is rooted to F. foetens (CBS 120665) and F. udum (CBS 177.31). Epi- and ex-type strains are indicated in bold.
