Abstract
Hepatocellular carcinoma (HCC) is a malignant tumor with high morbidity and mortality globally. It accounts for the majority of primary liver cancer cases. Amyloid precursor protein (APP), a cell membrane protein, plays a vital role in the pathogenesis of Alzheimer’s disease, and has been found to be implicated in tumor growth and metastasis. Therefore, to understand the relationship between APP and 5-fluorouracil (5-FU) resistance in liver cancer, Cell Counting Kit-8, apoptosis and cell cycle assays, western blotting, and reverse transcription-quantitative polymerase chain reaction (qPCR) analysis were performed. The results demonstrated that APP expression in Bel7402-5-FU cells was significantly up-regulated, as compared with that in Bel7402 cells. Through successful construction of APP-silenced (siAPP) and overexpressed (OE) Bel7402 cell lines, data revealed that the Bel7402-APP751-OE cell line was insensitive, while the Bel7402-siAPP cell line was sensitive to 5-FU in comparison to the matched control group. Furthermore, APP overexpression decreased, while APP silencing increased 5-FU-induced apoptosis in Bel7402 cells. Mechanistically, APP overexpression and silencing can regulate the mitochondrial apoptotic pathway and the expression of apoptotic suppressor genes (B-cell lymphoma-2 (Bcl-2) and B-cell lymphoma-extra large (Bcl-xl)). Taken together, these results preliminarily revealed that APP overexpression contributes to the resistance of liver cancer cells to 5-FU, providing a new perspective for drug resistance.
Keywords: Amyloid precursor protein, 5-Fluorouracil resistance, Mitochondrial apoptotic pathway, Hepatocellular carcinoma
1. Introduction
A liver cancer is a clinically challenging malignancy with high morbidity and mortality rates, of which hepatocellular carcinoma (HCC) accounts for most cases of liver cancer worldwide (Allemani et al., 2018). Recent cancer epidemiology statistics have indicated that hundreds of thousands of patients develop liver cancer in China every year (Siegel et al., 2018). Chemotherapy is a common treatment for liver cancer (Song, 2015; Ikeda et al., 2018), but the resistance of tumor cells limits its efficacy (Li et al., 2017; Niu et al., 2017). Drug resistance in hepatocarcinoma is a multi-step and multi-factor process, including a high expression level of multidrug resistance proteins (Ceballos et al., 2018), abnormal initiation of the apoptosis pathways (Li et al., 2018), and regulation of intracellular signaling pathways (Wang et al., 2018). However, existing research has not fully elucidated the mechanism of drug resistance in liver cancer. Therefore, further study of the underlying molecular mechanisms of drug resistance will help to identify novel drug targets.
Amyloid precursor protein (APP), a cell membrane protein, plays an important role in the pathogenesis of Alzheimer’s disease (Bukhari et al., 2017; Hardy, 2017). APP can be cleaved by β-and γ-secretase to produce β-amyloid (Aβ), the primary component of senile plaques. The soluble oligomer produced by APP exhibits neurotoxicity, which can inhibit the proliferation and differentiation of nerve cells; this can ultimately lead to cell dysfunction and apoptosis (Kallop et al., 2014; Rodríguez-Ruiz et al., 2017). This is the basis for one of the theories behind the etiology of Alzheimer’s disease. Previous studies have demonstrated that APP is expressed in certain neuronal stromal cells, and that it binds to death receptors on the surface of neuronal membranes through ectodomain fragments; this activates classical and non-canonical apoptotic pathways, inducing neuronal cell death, and promotes the occurrence and development of neurodegenerative diseases (Nikolaev et al., 2009; Kallop et al., 2014; Rodríguez-Ruiz et al., 2017).
In recent years, notably high expression levels of APP have been indicated in tumors, including breast (Harder et al., 2017; Gehrke et al., 2018), prostate (Takayama et al., 2009; Gough et al., 2014; Miyazaki et al., 2014), colorectal (Meng et al., 2001; Zhao et al., 2015), and pancreatic cancers (Hansel et al., 2003; Woods and Padmanabhan, 2013). Furthermore, elevated APP expression was found to negatively correlate with patient prognosis. Additional studies have shown that APP is involved in tumor hematogenous metastasis, and that cellular metastatic ability is significantly increased in tumor cells overexpressing APP (Strilic et al., 2016). In the present study, we found that APP expression was significantly up-regulated in 5-fluorouracil (5-FU)-resistant HCC cell lines relative to the parental cells, suggesting that APP may be involved in the drug-resistance process. Herein, the major aim of this study was to investigate the mechanism(s) responsible for APP-associated drug resistance in liver cancer cells.
2. Materials and methods
2.1. Cells
Human liver cancer Bel7402 and Bel7402-5-FU cell lines were obtained from and authenticated by the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (CAS, Shanghai, China), and maintained in our laboratory.
2.2. Reagents
Roswell Park Memorial Institute (RPMI) 1640 medium and fetal bovine serum (FBS) were purchased from Hyclone, GE Healthcare Life Sciences (Logan, UT, USA). Polyethylenimine (PEI) was purchased from Sangon Biotech Co., Ltd. (Shanghai, China), and 5-FU was purchased from Sigma-Aldrich, Merck KGaA (Darmstadt, Germany). Reverse transcription kits, Cell Counting Kit (CCK)-8 reagent and SYBR Green quantitative polymerase chain reaction (qPCR) kits were acquired from Beijing TransGen Biotech Co., Ltd. (Beijing, China). Rabbit anti-human APP (catalog No. 2452), BH3 interacting domain death agonist (BID; catalog No. 8762), B-cell lymphoma-2 (Bcl-2; catalog No. 4223), Bcl-2-associated X protein (BAX; catalog No. 5023), B-cell lymphoma-extra large (Bcl-xl; catalog No. 2764), c-Jun N-terminal kinase 3 (JNK3; catalog No. 2305), phosphorylated JNK3 (p-JNK3; catalog No. 8206), caspase-3 (catalog No. 9662), cleaved caspase-3 (catalog No. 9661), caspase-9 (catalog No. 9502), cleaved caspase-9 (catalog No. 9509), poly(ADP-ribose) polymerase 1 (PARP; catalog No. 9532), and cleaved PARP (catalog No. 9185), as well as mouse anti-human mixed-lineage kinase 3 (MLK3; catalog No. 2817) primary antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Rabbit anti-BID (catalog No. ab10640), p-MLK3 (catalog No. ab191530) and β-actin (catalog No. ab179467) primary antibodies, as well as horseradish peroxidase (HRP)-conjugated goat anti-rabbit (catalog No. ab6721) and HRP-conjugated goat anti-mouse (catalog No. ab6789) secondary antibodies were purchased from Abcam (Cambridge, UK). TRIzol® reagent was acquired from Thermo Fisher Scientific, Inc. (Waltham, MA, USA) and qPCR primers were synthesized by Sangon Biotech Co., Ltd. Polybrene was purchased from Tiangen Biotech Co., Ltd. (Beijing, China).
2.3. Cell culture
The human liver cancer Bel7402 and Bel7402-5-FU cells were cultured in RPMI 1640 medium supplemented with 10% (0.1 g/mL) FBS and 1% (0.01 g/mL) penicillin-streptomycin, at 37 °C in an incubator with 5% CO2.
2.4. CCK-8 assay
Human liver cancer cells were seeded into 96-well plates at a density of 1×104 cells per well and treated with 0.0 (control), 2.5, 5.0, 10.0, 20.0, 40.0, or 80.0 μg/mL 5-FU for 48 h. The supernatant was then discarded and 10% (0.1 g/mL) CCK-8 reagent was added, prior to a further 2-h incubation period. Absorbance values were measured at 450 nm using the iMark Microplate Reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The assay was conducted three times per group.
2.5. Plasmid construction and lentiviral infection
The human APP was synthesized and cloned into the multiple cloning sites (MCSs) of ampicillin-resistant plasmid pLV-CS (Shanghai Genechem Co., Ltd., China) to construct the pLV-CS-APP751-IRES (internal ribosome entry site)-GFP (green fluorescent protein) plasmid. A total of 1.5 μg pLV-CS-APP751-IRES-GFP plasmid and 1.5 μg packaging plasmid with ampicillin resistance (Shanghai Genechem Co., Ltd.) were added into 100 μL medium. Three packaging plasmids were added at a ratio of 5:3:2 (plasmid-encoded Gag-Pol precursor protein (pMDLg/pRRE): 0.75 μg; plasmid-encoded vesicular stomatitis virus G glycoprotein (pVSVG): 0.45 μg; plasmid-encoded regulator of expression of virion protein (pRSV-REV): 0.30 μg). Simultaneously, 6 μL PEI (10 μmol/L) was added and the solution was mixed, followed by incubation for 30 min at room temperature. Next, 900 μL complete culture medium was added to the mixed solution (PEI, 1 μmol/L). Subsequently, 1 mL of mixture was incubated with 293T cells. The medium was replaced after 12 h, and the supernatant was collected after 24 h. Subsequently, to construct the Bel7402-APP751-OE (APP overexpression) cell line, the supernatant was used for infection, where the supernatant was mixed with fresh medium at a ratio of 1:1 (v:v) and added into 293T cells. Polybrene was added to the cells at a final concentration of 10 μg/mL, followed by centrifugation at 37 °C, 400g for 30 min. Afterwards, they were incubated with 2 μg/mL of puromycin for 3 d to obtain the APP751-OE cell line. Additionally, the Bel7402-siAPP (APP-silenced) cell line was constructed using the pLVX-siAPP plasmid in the aforementioned manner.
2.6. Apoptosis and cycle cell assays
Human liver cancer Bel7402 and Bel7402-5-FU cells were seeded into a six-well plate at a density of 8.0×105 cells/well. Following adhesion, the supernatant was collected and the adherent cells were trypsinized; both were centrifuged at 250g for 3 min. After discarding the supernatant, the cells were fixed with pre-cooled ethanol (70% in phosphate-buffered saline (PBS)) at 4 °C overnight, and then harvested by centrifugation at 700g for 3 min. The cells were washed once and resuspended in 1 mL pre-cooled PBS, adjusted to a cell concentration of 1.0×106 cells/mL. Then RNase A enzyme was added, followed by propidium iodide (PI) staining for 10 min at room temperature. Subsequently, the cells were collected by centrifugation at 700g for 3 min, washed once, and resuspended in 1 mL pre-cooled PBS. Finally, the cells were filtered with a 75-micron nylon mesh before apoptosis and cell cycle analysis. The data were analyzed using FlowJo10 software (Tree Star, Inc., Ashland, OR, USA) and the experiments were conducted three times per group. In addition, apoptosis was detected using an Annexin V/PI staining kit (Beijing Zoman Biotechnology Co., Ltd., China) according to the manufacturer’s instructions, where the cells were treated as aforementioned, and detected by flow cytometry.
2.7. qPCR analysis
Total RNA was extracted from each group using TRIzol® reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer’s instructions. The purity and concentration of the total RNA were determined using the SmartSpec Plus Spectrophotometer (Bio-Rad, USA). A ratio of absorbance at 260 nm to that at 280 nm (A 260/A 280) in the range of 1.8–2.0 was considered acceptable. One microgram of total RNA was reverse-transcribed into complementary DNA (cDNA) using a reverse transcription kit according to the manufacturer’s protocol. Reverse transcription was performed at 37 °C for 15 min and 85 °C for 5 s, and the reactions were held at 4 °C. Subsequently, the cDNA was used as a template for qPCR. The reaction procedure was conducted using the ABI7500 qPCR instrument (Applied Biosystems, Thermo Fisher Scientific, Inc.) with the following thermocycling conditions: pre-denaturation at 95 °C for 5 min, denaturation at 95 °C for 15 s, and annealing at 60 °C for 15 s, for a total of 40 cycles. The relative quantitative expression of target genes was normalized to that of β-actin and calculated using the 2−ΔΔ C T method. All experiments were conducted three times. The primer sequences were as follows: APP forward, TCTCGTTCCTGACAAGTGCAA and reverse, GCAAGTTGGTACTCTTCTCACTG; caspase-3 forward, CATGGAAGCGAATCAATGGACT and reverse, CTGTACCAGACCGAGATGTCA; caspase-9 forward, CCTGGAGCGGATTACCCCT and reverse, CTGTATGCTGGTGTCTAGGAGA; BAX forward, CCCGAGAGGTCTTTTTCCGAG and reverse, CC AGCCCATGATGGTTCTGAT; BID forward, ATG GACCGTAGCATCCCTCC and reverse, GTAGGT GCGTAGGTTCTGGT; Bcl-2 forward, GGTGGGG TCATGTGTGTGG and reverse, CGGTTCAGGTAC TCAGTCATCC; Bcl-xl forward, GAGCTGGTGGTT GACTTTCTC and reverse, TCCATCTCCGATTCAG TCCCT; MLK3 forward, AAGGAAAAGGAACTA CTGAGCCG and reverse, GCTCGAACACCTCTA GCTCC; JNK3 forward, TATGTGGAGAATCGGC CCAAG and reverse, GCTTTGAGTTTATTGTGC TCGG; β-actin forward, CCTCGCCTTTGCCGAT CC and reverse, GGATCTTCATGAGGTAGTCAGTC.
2.8. Western blot analysis
Liver cancer cells were seeded into a six-well plate at a density of 5.0×105 cells per well. The cells were then treated with 20 μg/mL 5-FU for 24 h, and the total protein was extracted from each group using radioimmunoprecipitation assay (RIPA) buffer, prior to quantification using the bicinchoninic acid (BCA) method. Proteins were isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 10% (0.1 g/mL) gel, and transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were then blocked with 5% bovine serum albumin (BSA; Solarbio, Beijing, China) for 2 h at room temperature, and subsequently incubated with the corresponding primary antibodies overnight at 4 °C. After washing with Tris-buffered saline with Tween 20 (TBST), the secondary antibodies were added and the membranes were incubated for 2 h at room temperature. The bands were developed with chemiluminescence reagents (EMD Millipore) and imaged using the ImageQuant LAS 4000 mini (GE Healthcare Life Sciences). The experiments were repeated three times.
2.9. Statistical analysis
SPSS 21.0 (IBM Corp., Armonk, NY, USA) was used to perform all statistical data analyses. The two-tailed Student’s t-test was used to compare the differences between two groups, and one-way analysis of variance (ANOVA) with least significant difference (LSD) test was performed to analyze those between multiple groups. The data are presented as the mean±standard deviation (SD), and P<0.05 was considered to indicate a statistically significant difference.
3. Results
3.1. Expression of APP in Bel7402-5-FU HCC cells
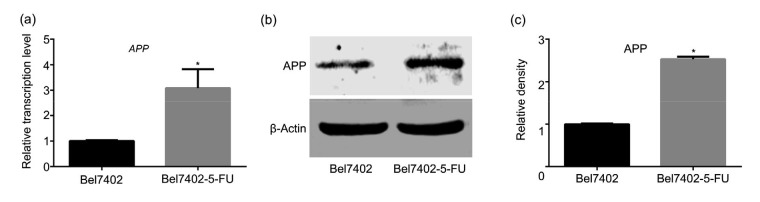
To understand the mechanism of APP in the 5-FU-resistance of liver cancer cells, the expression level of APP was determined by qPCR and western blotting. The results showed that compared with Bel7402 cells, the APP expression in Bel7402-5-FU cells was significantly up-regulated at both the transcriptional and translational levels (P<0.05; Fig. 1).
Fig. 1.
Significantly up-regulated expression of APP in Bel7402-5-FU HCC cells
(a) mRNA expression levels of APP in Bel7402 and Bel7402-5-FU cells. (b, c) Protein expression levels of APP in Bel7402-5-FU cells were higher than those in Bel7402 cells. (a, c) Data are expressed as mean±SD (n=3). * P<0.05, compared with the control group. APP: amyloid precursor protein; 5-FU: 5-fluorouracil
3.2. Successful construction of APP-silenced and -overexpressed Bel7402 cell lines
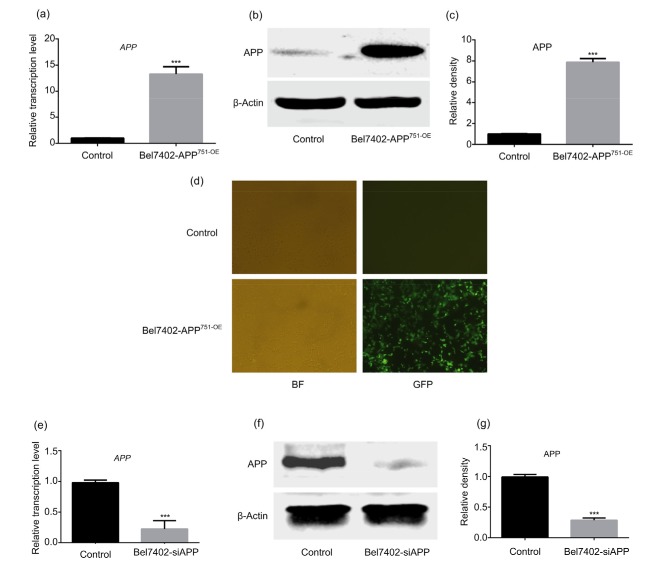
Virus infection efficiency was validated by qPCR, western blotting, and immunofluorescence analysis. As presented in Fig. 2, APP overexpression (Figs. 2a–2d) and silencing were observed at both the transcriptional and translational levels, compared with the matched control group (Figs. 2e–2g; P<0.05), suggesting that Bel7402-APP751-OE and Bel7402-siAPP cell lines were successfully constructed.
Fig. 2.
Successful construction of Bel7402-APP751-OE and Bel7402-siAPP cell lines
(a) Statistical analysis of the mRNA expression levels of APP in Bel7402-APP751-OE cells. (b, c) Representative western blot analysis of APP protein expression in Bel7402-APP751-OE cells. (d) Representative immunofluorescence images of GFP expression in Bel7402-APP751-OE cells. (e) Statistical analysis of the mRNA expression levels of APP in Bel7402-siAPP cells. (f, g) Representative western blot analysis of APP protein expression in Bel7402-siAPP cells. (a, c, e, g) Data are expressed as mean±SD (n=3). *** P<0.001, compared with the control group. BF: bright field; GFP: green fluorescent protein; APP: amyloid precursor protein; OE: overexpression; siAPP: silenced APP
3.3. Effect of 5-FU on Bel7402-APP751-OE and Bel7402-siAPP cell lines
Cells were treated with various concentrations of 5-FU (0.0, 2.5, 5.0, 10.0, 20.0, 40.0, and 80.0 μg/mL) for 48 h, and the absorbance value was determined after incubation with CCK-8 reagent. Fig. 3 indicates that 5-FU treatment significantly inhibited the proliferation of Bel7402 cells (P<0.05), but no obvious difference was observed in Bel7402-APP751-OE cells, compared with that in Bel7402 cells (Fig. 3a). Moreover, the half maximal inhibitory concentration (IC50) of 5-FU in Bel7402-APP751-OE cells was markedly higher than that in the Bel7402 cells (Fig. 3b; P<0.01). By contrast, when compared with the control group, the opposite effect was observed in Bel7402-siAPP cells (Figs. 3c and 3d).
Fig. 3.
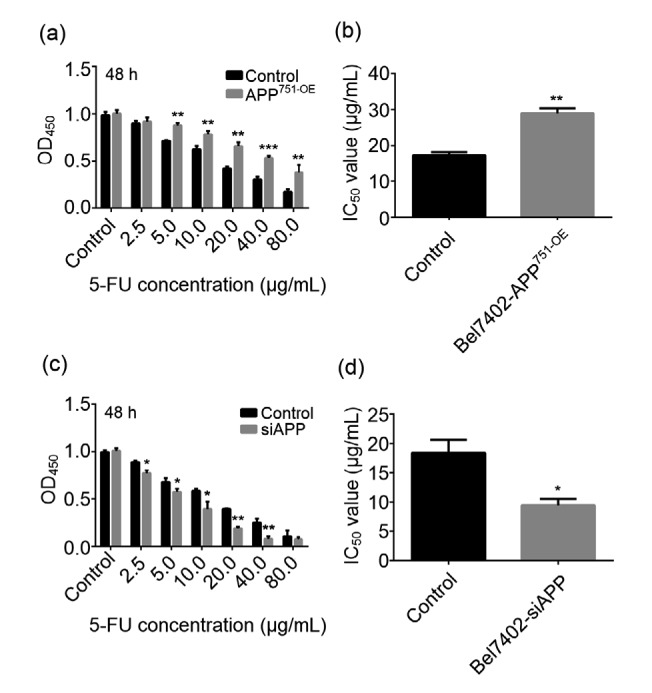
Opposite effect of 5-FU treatment on Bel7402-APP751-OE and Bel7402-siAPP cell lines
(a) Statistical analyses of 5-FU cytotoxicities in Bel7402 and Bel7402-APP751-OE cells using the CCK-8 assay. (b) IC50 value of 5-FU in Bel7402 and Bel7402-APP751-OE cells. (c) Statistical analyses of 5-FU cytotoxicities in Bel7402 and Bel7402-siAPP cells using the CCK-8 assay. (d) IC50 value of 5-FU in Bel7402 and Bel7402-siAPP cells. Data are expressed as mean±SD (n=3). * P<0.05 and ** P<0.01, compared with the control group. APP: amyloid precursor protein; CCK-8: Cell Counting Kit-8; 5-FU: 5-fluorouracil; OD450: optical density value in 450 nm; IC50: half maximal inhibitory concentration; OE: overexpression; siAPP: silenced APP
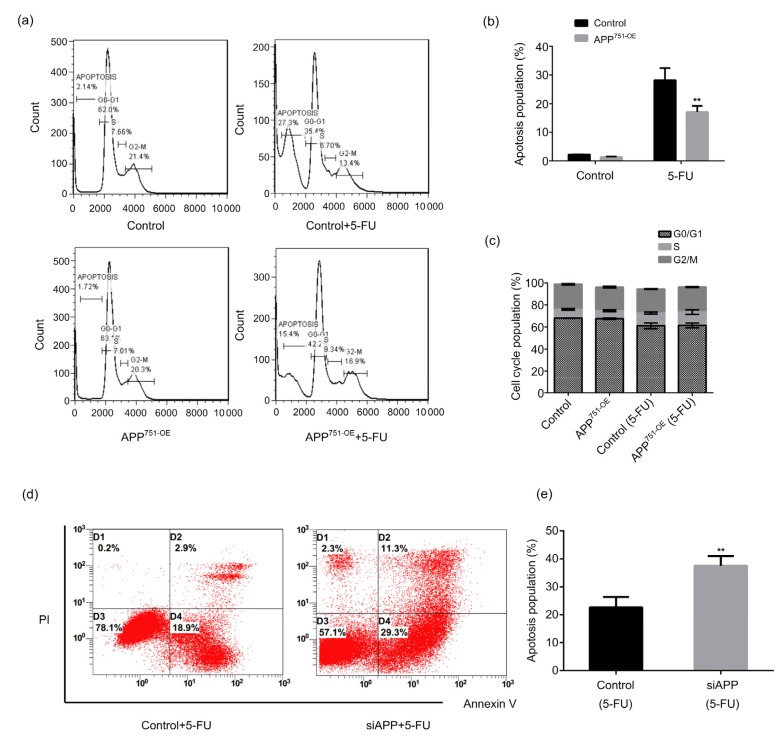
3.4. Effect of APP on 5-FU-induced apoptosis in Bel7402 cells
The results in Fig. 4 reveal that 5-FU-induced apoptosis in Bel7402-APP751-OE cells was significantly lower than that in Bel7402 cells (Figs. 4a and 4b; P<0.05), though no significant effect on the cell cycle was observed in either cell type (Fig. 4c; P>0.05). By contrast, 5-FU-induced apoptosis was significantly increased in Bel7402-siAPP cells compared with the control group (Figs. 4d and 4e; P<0.05).
Fig. 4.
Effect of APP on 5-FU-induced apoptosis in Bel7402 cells
(a, b) 5-FU (40 μg/mL) induced apoptosis in Bel7402 and Bel7402-APP751-OE cells, as assessed by flow cytometry. (c) Statistical analysis of the cell cycle in Bel7402 and Bel7402-APP751-OE cells following treatment with 5-FU (40 μg/mL). (d, e) 5-FU (40 μg/mL) induced apoptosis in Bel7402 and Bel7402-siAPP cells, as determined by Annexin V/propidium iodide staining. (b, c, e) Data are expressed as mean±SD (n=3). ** P<0.01, compared with the control. APP: amyloid precursor protein; 5-FU: 5-fluorouracil; OE: overexpression; siAPP: silenced APP; PI: propidium iodide
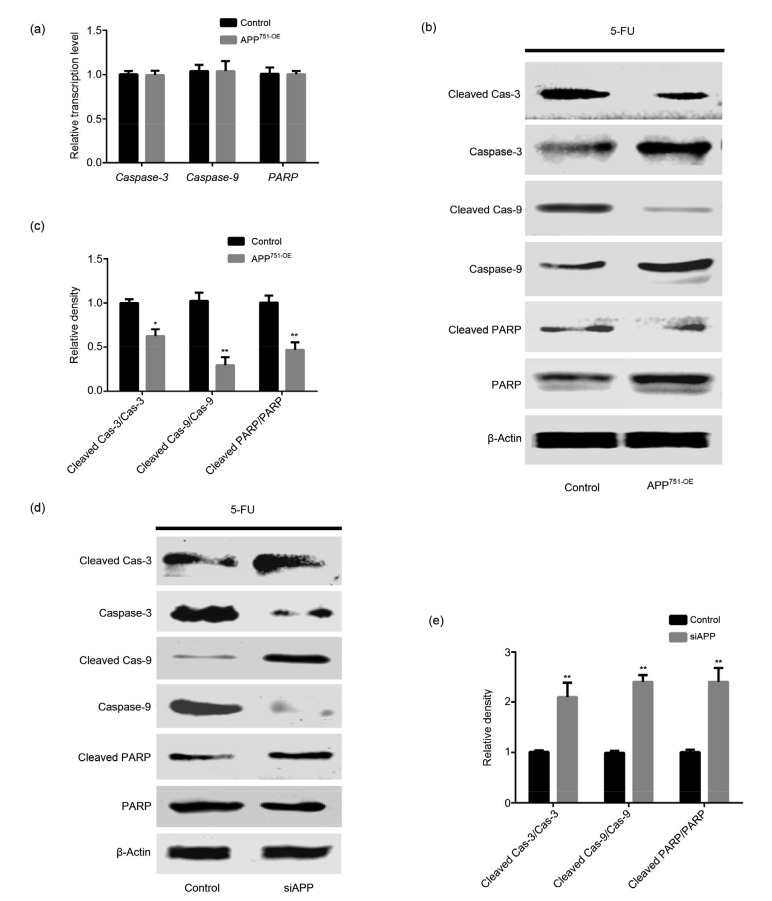
3.5. Effect of APP on the mitochondrial apoptotic pathway
Bel7402, Bel7402-siAPP, and Bel7402-APP751-OE cells were cultured in 6-well plates, and incubated with 40 μg/mL of 5-FU. The expression levels and activation of caspase family-related molecules were determined. The results suggested that when treated with 5-FU, the activation of caspase-3, caspase-9, and their substrate PARP were markedly attenuated in Bel7402-APP751-OE cells compared with Bel7402 cells (Fig. 5c; P<0.05); no significant difference in the mRNA or protein expression level was observed in either cell line (Figs. 5a and 5b; P>0.05). Conversely, the activation of caspase-3, caspase-9, and PARP was markedly up-regulated in Bel7402-siAPP cells compared with that in Bel7402 cells (Figs. 5d and 5e; P<0.05).
Fig. 5.
Effects of APP on the activation of the caspase family proteins and PARP
(a) mRNA expression levels of caspase-3, caspase-9, and PARP were detected by qPCR, and no statistical differences were revealed between the two groups. (b, c) Protein expression levels of caspase-3, caspase-9, PARP, cleaved caspase-3, cleaved caspase-9, and cleaved PARP were analyzed by western blot assay, of which protein expression levels of cleaved caspase-3, cleaved caspase-9, and cleaved PARP were significantly down-regulated in Bel7402-APP751-OE cells, while caspase-3, caspase-9, and PARP expression levels were greatly up-regulated, as compared to those in Bel7402 cells. (d, e) Protein expression levels of cleaved caspase-3, cleaved caspase-9, and cleaved PARP were significantly increased in Bel7402-siAPP cells, whereas protein expression levels of caspase-3, caspase-9, and PARP were dramatically decreased, as compared with those in Bel7402 cells. (a, c, e) Data are presented as the mean±SD of three experiments with replicates. * P<0.05 and ** P<0.01, compared with the control. APP: amyloid precursor protein; OE: overexpression; siAPP: silenced APP; PARP: poly(ADP-ribose) polymerase 1; 5-FU: 5-fluorouracil
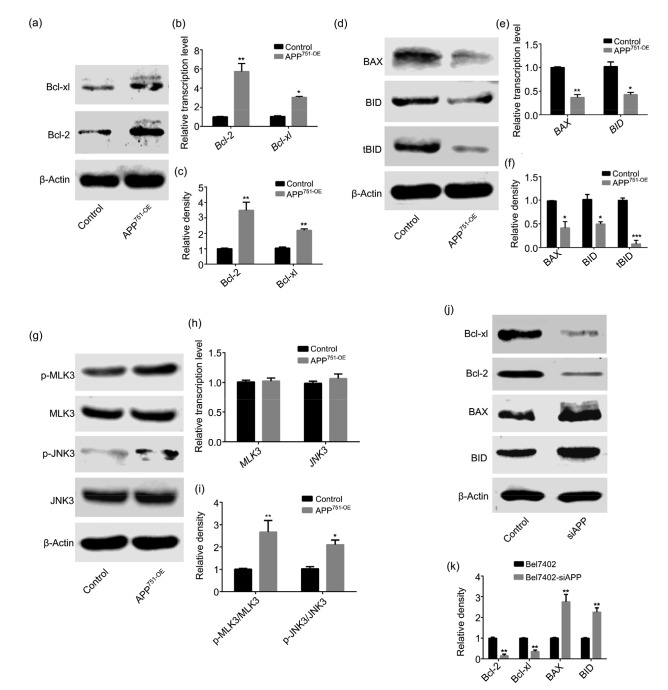
3.6. Effect of APP on the expression of apoptotic suppressor genes
The results in Fig. 6 show that compared with those in Bel7402 cells, the mRNA and protein expression levels of mitochondrial pathway-related molecules BID and BAX were significantly down-regulated in Bel7402-APP751-OE cells (Figs. 6d–6f; P<0.05), while those of the survival-related genes Bcl-2 and Bcl-xl were significantly up-regulated (Figs. 6a–6c; P<0.05). Moreover, activation of mitogen-activated protein kinase (MAPK) pathway-related proteins MLK3 and JNK3 was highly elevated when compared with the control group (Fig. 6i; P<0.05), even though the mRNA or protein expression level did not change significantly (Figs. 6g and 6h; P>0.05). Rather, western blotting revealed that the protein expression of Bcl-2 and Bcl-xl was remarkably attenuated after APP silencing as compared with the control group (Figs. 6j and 6k; P<0.05).
Fig. 6.
Effects of APP on the expression of mitochondrial pathway-and MAPK pathway-related molecules
(a, c) Protein expression levels of Bcl-2 and Bcl-xl were significantly up-regulated in Bel7402-APP751-OE cells, compared with those in Bel7402 cells. (b) mRNA expression levels of Bcl-2 and Bcl-xl were markedly increased in Bel7402-APP751-OE cells, compared with those in Bel7402 cells. (d, f) Protein expression levels of BAX and BID were significantly down-regulated in Bel7402-APP751-OE cells, compared with those in Bel7402 cells. (e) mRNA expression levels of BAX and BID were greatly decreased in Bel7402-APP751-OE cells in comparison to those in Bel7402 cells. (g, h) No significant differences were observed in the protein or mRNA expression levels of MLK3 and JNK3 in either Bel7402-APP751-OE or Bel7402 cells. (i) Phosphorylation of MLK3 and JNK3 was elevated in Bel7402-APP751-OE cells, compared with that in Bel7402 cells. (j, k) Protein expression levels of Bcl-2 and Bcl-xl were markedly attenuated, but those of BAX and BID were up-regulated after APP silencing, compared with the control group. Data are presented as the mean±SD of three experiments with replicates. * P<0.05, ** P<0.01, and *** P<0.001, compared with the control. APP: amyloid precursor protein; MAPK: mitogen-activated protein kinase; MLK3: mixed-lineage kinase 3; JNK3: c-Jun N-terminal kinase 3; p-: phosphorylated; OE: overexpression; siAPP: silenced APP; BAX: B-cell lymphoma assosiated X; BID: BH3 interacting domain death agonist; tBID: total BID
4. Discussion
In this study, our findings revealed that APP expression was significantly up-regulated in 5-FU-resistant liver cancer cells. Moreover, an attenuation in 5-FU toxicity was observed in liver cancer cells following APP overexpression. In addition, APP overexpression inhibited the activation of apoptosis pathways, thereby inhibiting 5-FU-induced apoptosis in HCC cells, which was related to the regulation of the mitochondrial apoptotic pathway. In contrast to what was observed in Bel7402-APP751-OE cells, silencing APP in Bel7402 cells led to opposite results. Targeting APP may be a new perspective for drug resistance in liver cancer.
Drug resistance seriously restricts the success rate of liver cancer treatment, and subsequently affects patient prognosis (Hirata et al., 2015; Liu et al., 2016; Sun et al., 2017; Zhang et al., 2017). Studies have shown that liver cancer resistance is associated with a variety of factors (Hirata et al., 2015; Zhang et al., 2017). Thus, investigating the mechanisms of drug resistance and identifying new drug resistance-related genes are still of great clinical value. In the present study, we found that the expression of APP in human multidrug-resistant HCC cell lines was significantly higher than that in the parental cells, suggesting that APP may play a role in the drug-resistance process of liver cancer.
In order to understand whether APP plays a regulatory role in liver cancer-associated drug resistance, we successfully constructed the Bel7402-APP751-OE and Bel7402-siAPP cell lines, and confirmed that the sensitivity of Bel7402-APP751-OE cells to 5-FU was obviously reduced. Moreover, the cytotoxic effects of 5-FU on Bel7402 cells were stronger than those on Bel7402-APP751-OE cells; the IC50 values of 5-FU in Bel7402 cells were lower than those in Bel7402-APP751-OE cells, suggesting that APP overexpression contributes to liver cancer-associated 5-FU resistance. By contrast, data from the Bel7402-siAPP cell line further validated the role of APP in liver cancer cells.
5-FU causes apoptosis by inhibiting DNA and RNA syntheses (Zha et al., 2014; Dai et al., 2016). When treated with 5-FU, the apoptotic rates of both cell lines were significantly increased, but the apoptotic rate of Bel7402-APP751-OE cells was significantly lower than that of Bel7402 cells. In addition, no significant difference was observed in the cell cycle, indicating that APP overexpression specifically inhibits 5-FU-induced apoptosis. This is consistent with the results of the cell proliferation assay. Furthermore, previous studies have shown that increasing apoptosis may reduce cellular drug resistance (Ma et al., 2016; Guo et al., 2017), and thus combined with our results, this demonstrates that APP inhibits apoptosis, resulting in increased drug resistance.
Caspase family members play a vital role in apoptosis (Belmokhtar et al., 2001; Shi, 2002). In the present study, we found that 5-FU induced apoptosis, in which the activation of caspase-3, caspase-9, and their substrate PARP was significantly more down-regulated in Bel7402-APP751-OE cells than in Bel7402 cells; however, the expression levels of these proteins were not affected. In addition, the mitochondrial apoptotic pathway has a very important regulatory role in apoptosis (Wang et al., 2017). In this study, compared with Bel7402 cells, the expression of apoptosis-related proteins BAX and BID was significantly down-regulated in Bel7402-APP751-OE cells, while the expression of apoptosis inhibitory proteins Bcl-2 and Bcl-xl was significantly up-regulated. This was contrary to the results in Bel7402-siAPP cells, suggesting that APP may be regulated by the mitochondrial pathway. MLK3 and JNK3 are critical components of the MAPK signaling pathway, and may be downstream targets of APP (Kovalenko et al., 2012; Gao et al., 2019). Our data revealed that the expression levels of MLK3 and JNK3 did not change significantly, but that p-JNK3 and p-MLK3 levels were significantly up-regulated. Therefore, APP overexpression was activated by the MAPK signaling pathway. This was likely to involve the inhibition of apoptosis. However, the exact mechanisms require further elucidation.
In conclusion, APP may activate the MAPK signaling pathway, thereby regulating the expression of mitochondrial pathway-related proteins, Bcl-2, Bcl-xl, BAX, and BID, and modulating the resistance of liver cancer cells to chemotherapeutic drugs. Therefore, the activity of APP provides a novel insight for overcoming drug resistance in liver cancer.
Contributors
Study concept and design: Zhong-quan ZHAO. Performing the experiments: Xiao-long WU, Ying CHEN, and Wen-cui KONG. Experimental technical guidance: Zhong-quan ZHAO. Data analysis and manuscript drafting: Xiao-long WU and Zhong-quan ZHAO. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. All authors have read and approved the final manuscript. Therefore, all authors have full access to all the data in the study and take responsibility for the integrity and security of the data
Footnotes
Project supported by the International Science & Technology Cooperation Program of China (No. 2016G02)
Contributors: Study concept and design: Zhong-quan ZHAO. Performing the experiments: Xiao-long WU, Ying CHEN, and Wen-cui KONG. Experimental technical guidance: Zhong-quan ZHAO. Data analysis and manuscript drafting: Xiao-long WU and Zhong-quan ZHAO. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. All authors have read and approved the final manuscript. Therefore, all authors have full access to all the data in the study and take responsibility for the integrity and security of the data
Compliance with ethics guidelines: Xiao-long WU, Ying CHEN, Wen-cui KONG, and Zhong-quan ZHAO declare that there are no conflicts of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Allemani C, Matsuda T, di Carlo V, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/s0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belmokhtar CA, Hillion J, Ségal-Bendirdjian E. Staurosporine induces apoptosis through both caspase-dependent and caspase-independent mechanisms. Oncogene. 2001;20(26):3354–3362. doi: 10.1038/sj.onc.1204436. [DOI] [PubMed] [Google Scholar]
- 3.Bukhari H, Glotzbach A, Kolbe K, et al. Small things matter: implications of APP intracellular domain AICD nuclear signaling in the progression and pathogenesis of Alzheimer’s disease. Prog Neurobiol. 2017;156:189–213. doi: 10.1016/j.pneurobio.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Ceballos MP, Decándido G, Quiroga AD, et al. Inhibition of sirtuins 1 and 2 impairs cell survival and migration and modulates the expression of P-glycoprotein and MRP3 in hepatocellular carcinoma cell lines. Toxicol Lett. 2018;289:63–74. doi: 10.1016/j.toxlet.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Dai W, Gao QG, Qiu JP, et al. Quercetin induces apoptosis and enhances 5-FU therapeutic efficacy in hepatocellular carcinoma. Tumor Biol. 2016;37(5):6307–6313. doi: 10.1007/s13277-015-4501-0. [DOI] [PubMed] [Google Scholar]
- 6.Gao SY, Lin J, Wang T, et al. Qingxin Kaiqiao Fang ameliorates memory impairment and inhibits apoptosis in APP/PS1 double transgenic mice through the MAPK pathway. Drug Des Devel Ther. 2019;13:459–475. doi: 10.2147/dddt.s188505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gehrke A, Lee SS, Hilton K, et al. Development of the cancer survivor profile-breast cancer (CSPro-BC) app: patient and nurse perspectives on a new navigation tool. J Cancer Surviv. 2018;12(3):291–305. doi: 10.1007/s11764-017-0668-2. [DOI] [PubMed] [Google Scholar]
- 8.Gough M, Blanthorn-Hazell S, Delury C, et al. The E1 copper binding domain of full-length amyloid precursor protein mitigates copper-induced growth inhibition in brain metastatic prostate cancer DU145 cells. Biochem Biophys Res Commun. 2014;453(4):741–747. doi: 10.1016/j.bbrc.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo YK, Shi M, Qin YL, et al. Mechanism of apoptosis for resveratrol-mediated reversing the drug-resistance of AML HL-60/ADR cells. J Exp Hematol. 2017;25(3):736–742. doi: 10.7534/j.issn.1009-2137.2017.03.018. (in Chinese) [DOI] [PubMed] [Google Scholar]
- 10.Hansel DE, Rahman A, Wehner S, et al. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003;63(21):7032–7037. [PubMed] [Google Scholar]
- 11.Harder H, Holroyd P, Burkinshaw L, et al. A user-centred approach to developing bWell, a mobile app for arm and shoulder exercises after breast cancer treatment. J Cancer Surviv. 2017;11(6):732–742. doi: 10.1007/s11764-017-0630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy J. The discovery of Alzheimer-causing mutations in the APP gene and the formulation of the “amyloid cascade hypothesis”. FEBS J. 2017;284(7):1040–1044. doi: 10.1111/febs.14004. [DOI] [PubMed] [Google Scholar]
- 13.Hirata H, Sugimachi K, Takahashi Y, et al. Downregulation of PRRX1 confers cancer stem cell-like properties and predicts poor prognosis in hepatocellular carcinoma. Ann Surg Oncol. 2015;22(S3):1402–1409. doi: 10.1245/s10434-014-4242-0. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda M, Morizane C, Ueno M, et al. Chemotherapy for hepatocellular carcinoma: current status and future perspectives. Jpn J Clin Oncol. 2018;48(2):103–114. doi: 10.1093/jjco/hyx180. [DOI] [PubMed] [Google Scholar]
- 15.Kallop DY, Meilandt WJ, Gogineni A, et al. A death receptor 6-amyloid precursor protein pathway regulates synapse density in the mature CNS but does not contribute to Alzheimer’s disease-related pathophysiology in murine models. J Neurosci. 2014;34(19):6425–6437. doi: 10.1523/jneurosci.4963-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovalenko PL, Kunovska L, Chen J, et al. Loss of MLK3 signaling impedes ulcer healing by modulating MAPK signaling in mouse intestinal mucosa. Am J Physiol Gastrointest Liver Physiol. 2012;303(8):G951–G960. doi: 10.1152/ajpgi.00158.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Duan BJ, Guo Y, et al. Baicalein sensitizes hepatocellular carcinoma cells to 5-FU and epirubicin by activating apoptosis and ameliorating P-glycoprotein activity. Biomed Pharmacother. 2018;98:806–812. doi: 10.1016/j.biopha.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Li JQ, Wu X, Gan L, et al. Hypoxia induces universal but differential drug resistance and impairs anticancer mechanisms of 5-fluorouracil in hepatoma cells. Acta Pharmacol Sin. 2017;38(12):1642–1654. doi: 10.1038/aps.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu JX, Cui XP, Qu LS, et al. Overexpression of DLX2 is associated with poor prognosis and sorafenib resistance in hepatocellular carcinoma. Exp Mol Pathol. 2016;101(1):58–65. doi: 10.1016/j.yexmp.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Ma ST, Tan WH, Du BT, et al. Oridonin effectively reverses cisplatin drug resistance in human ovarian cancer cells via induction of cell apoptosis and inhibition of matrix metalloproteinase expression. Mol Med Rep. 2016;13(4):3342–3348. doi: 10.3892/mmr.2016.4897. [DOI] [PubMed] [Google Scholar]
- 21.Meng JY, Kataoka H, Itoh H, et al. Amyloid β protein precursor is involved in the growth of human colon carcinoma cell in vitro and in vivo . Int J Cancer. 2001;92(1):31–39. doi: 10.1002/1097-0215(200102)9999:9999<::AID-IJC1155>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki T, Ikeda K, Horie-Inoue K, et al. Amyloid precursor protein regulates migration and metalloproteinase gene expression in prostate cancer cells. Biochem Biophys Res Commun. 2014;452(3):828–833. doi: 10.1016/j.bbrc.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Nikolaev A, McLaughlin T, O'Leary DDM, et al. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457(7232):981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Niu LL, Liu LP, Yang SL, et al. New insights into sorafenib resistance in hepatocellular carcinoma: responsible mechanisms and promising strategies. Biochim Biophys Acta Rev Cancer. 2017;1868(2):564–570. doi: 10.1016/j.bbcan.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Rodríguez-Ruiz M, Moreno E, Moreno-Delgado D, et al. Heteroreceptor complexes formed by dopamine D1, histamine H3, and N-methyl-D-aspartate glutamate receptors as targets to prevent neuronal death in Alzheimer’s disease. Mol Neurobiol. 2017;54(6):4537–4550. doi: 10.1007/s12035-016-9995-y. [DOI] [PubMed] [Google Scholar]
- 26.Shi YG. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9(3):459–470. doi: 10.1016/S1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 27.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 28.Song MJ. Hepatic artery infusion chemotherapy for advanced hepatocellular carcinoma. World J Gastroenterol. 2015;21(13):3843–3849. doi: 10.3748/wjg.v21.i13.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strilic B, Yang LD, Albarrán-Juárez J, et al. Tumour-cell-induced endothelial cell necroptosis via death receptor 6 promotes metastasis. Nature. 2016;536(7615):215–218. doi: 10.1038/nature19076. [DOI] [PubMed] [Google Scholar]
- 30.Sun T, Liu H, Ming L. Multiple roles of autophagy in the sorafenib resistance of hepatocellular carcinoma. Cell Physiol Biochem. 2017;44(2):716–727. doi: 10.1159/000485285. [DOI] [PubMed] [Google Scholar]
- 31.Takayama K, Tsutsumi S, Suzuki T, et al. Amyloid precursor protein is a primary androgen target gene that promotes prostate cancer growth. Cancer Res. 2009;69(1):137–142. doi: 10.1158/0008-5472.can-08-3633. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Xu B, Li QX, et al. Anti-cancer effects of a novel Pan-RAF inhibitor in a hepatocellular carcinoma cell line. Mol Med Rep. 2018;17(4):6185–6193. doi: 10.3892/mmr.2018.8615. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Lu XC, Zhu RL, et al. Betulinic acid induces apoptosis in differentiated PC12 cells via ROS-mediated mitochondrial pathway. Neurochem Res. 2017;42(4):1130–1140. doi: 10.1007/s11064-016-2147-y. [DOI] [PubMed] [Google Scholar]
- 34.Woods NK, Padmanabhan J. Inhibition of amyloid precursor protein processing enhances gemcitabine-mediated cytotoxicity in pancreatic cancer cells. J Biol Chem. 2013;288(42):30114–30124. doi: 10.1074/jbc.M113.459255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zha Y, Gan P, Yao Q, et al. Downregulation of Rap1 promotes 5-fluorouracil-induced apoptosis in hepatocellular carcinoma cell line HepG2. Oncol Rep. 2014;31(4):1691–1698. doi: 10.3892/or.2014.3033. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Luo N, Tian Y, et al. USP22 knockdown enhanced chemosensitivity of hepatocellular carcinoma cells to 5-FU by up-regulation of Smad4 and suppression of Akt. Oncotarget. 2017;8(15):24728–24740. doi: 10.18632/oncotarget.15798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao YJ, Han HZ, Liang Y, et al. Alternative splicing of VEGFA, APP and NUMB genes in colorectal cancer. World J Gastroenterol. 2015;21(21):6550–6560. doi: 10.3748/wjg.v21.i21.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]