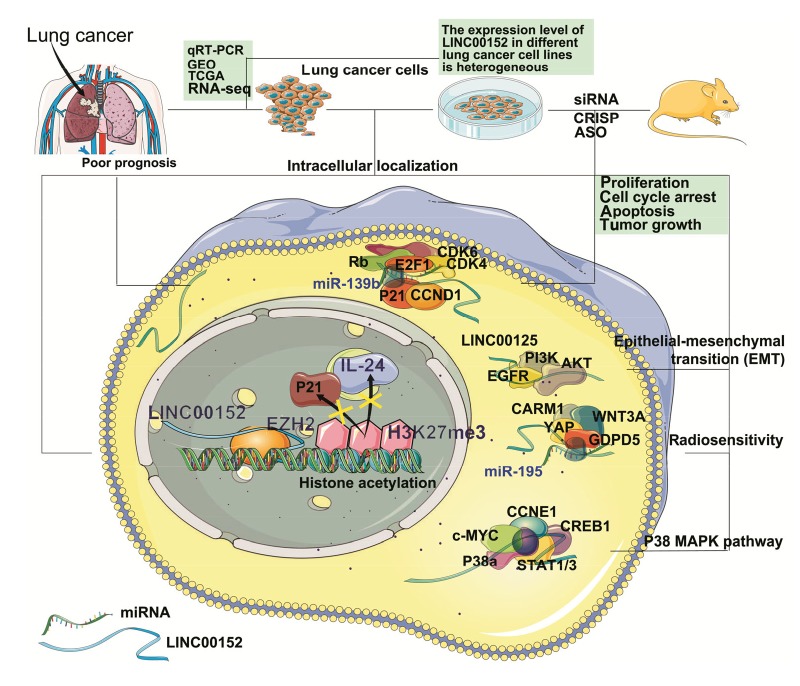
Fig. 1.
Potential mechanisms of LINC00152 in NSCLC
LINC00152 has been associated with poor prognosis of non-small cell lung cancer (NSCLC) patients. It has been shown to be highly expressed in tumor tissues and peripheral blood samples of NSCLC patients through analysis of the data from quantitative real-time polymerase chain reaction (qRT-PCR) assay, RNA sequencing (RNA-seq), The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. In vitro, the expression level of LINC00152 in different NSCLC cell lines is heterogenous. It is located in both cytoplasm and the nucleus, and it can exert carcinogenic effects through different molecular mechanisms. In the nucleus, LINC00152 regulates gene expression by binding to enhancer of zeste homolog 2 (EZH2), while it affects cellular signaling cascades in the cytoplasm. It can also bind specific microRNAs (miRNAs) as competing endogenous RNA (ceRNA) to regulate the expression of target messenger RNAs (mRNAs). On the other hand, the mechanisms of LINC00152 can be studied in vivo by regulating its expression using small interfering RNA (siRNA), clustered regularly interspaced short palindromic repeat (CRISPR), and antisense oligonucleotide (ASO) approaches in xenograft models. CDK: cyclin-dependent kinase; E2F1: E2F transcription factor 1; CCND1: cyclin D1; PI3K: phosphoinositide-3-kinase; EGFR: epidermal growth factor receptor; CARM1: coactivator-associated arginine methyltransferase 1; WNT3A: Wnt family member 3A; YAP: yes-associated protein 1; GDPD5: glycerophosphodiester phosphodiesterase domain containing 5; CCNE1: cyclin E1; CREB1: cyclic adenosine monophosphate (cAMP)-responsive element binding protein 1; STAT1/3: signal transducer and activator of transcription 1/3