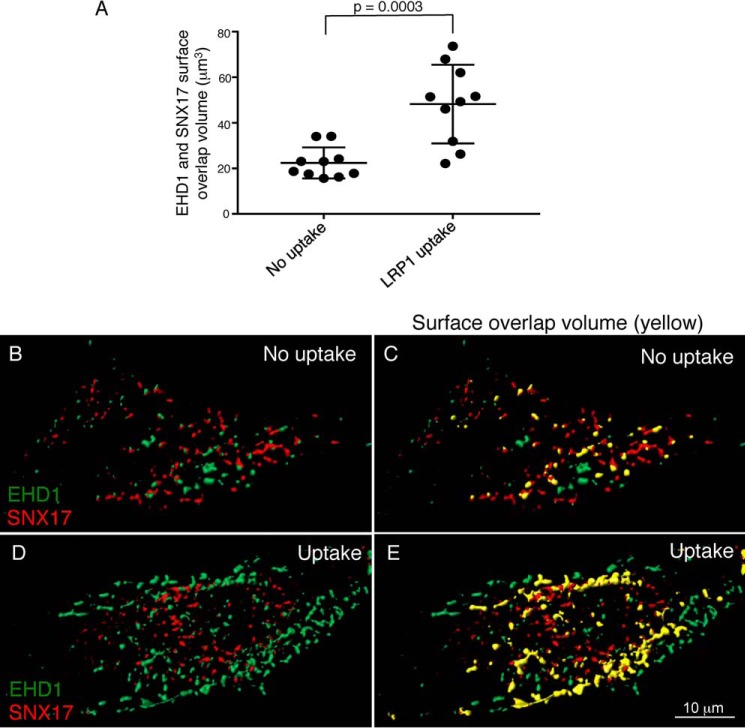
Figure 4.
EHD1 and SNX17 surface overlap volume increases upon LRP1 uptake. CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1-GFP were mock-treated (no uptake) or incubated with anti-LRP1 antibody as described in Fig. 3. The cells were fixed and stained with anti-SNX17 antibody and imaged by confocal microscopy. Z-stacks were acquired and processed by IMARIS. 3D surface reconstruction was performed simultaneously for both channels to capture EHD1 and SNX17 voxels. Surface–surface overlap volume was assessed using the IMARIS XT bundle Kiss and Run by integrating it with MATLAB Compiler Runtime and launching on IMARIS. A, EHD1 surfaces were selected as target surfaces, and SNX17 surfaces were tracked for any overlapping voxels with those of EHD1. The total surface overlap volume between EHD1 and SNX17 surfaces was quantified and plotted for the no uptake and LRP1 uptake conditions. Two-tailed t tests were performed. B–E, representative images showing 3D surface reconstruction for EHD1 and SNX17 surfaces without (B and C) and with LRP1 uptake (D and E). The overlap is indicated in yellow (C and E). Data shown are representative of three independent experiments.