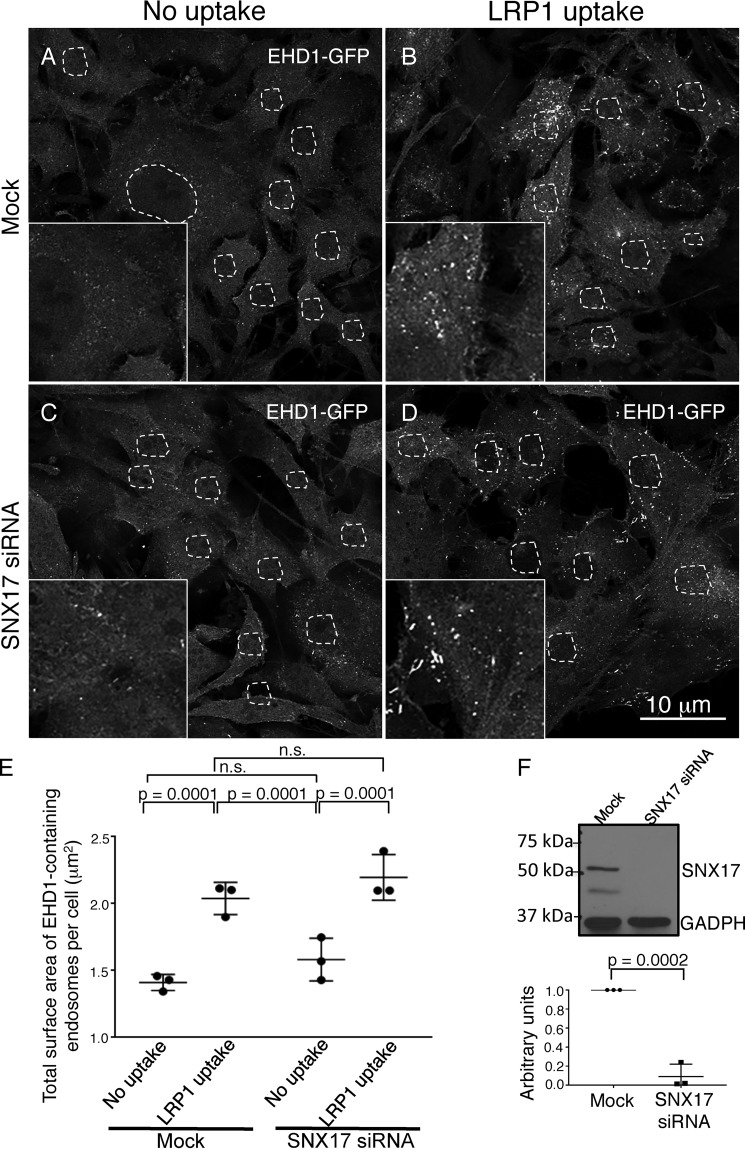
Figure 5.
EHD1 is recruited to endosomes in the absence of SNX17. A–D, CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1-GFP were mock-treated (A and B) or subjected to SNX17 siRNA (C and D) and incubated with LRP1 antibodies (B and D) or left untreated as a control (A and C). E, z-sections obtained from confocal microscopy were processed with IMARIS software to construct 3D surfaces for EHD1, as discussed under “Experimental procedures,” and the total EHD1 surface area per cell was calculated. The graph depicts the total surface area of EHD1-containing endosomes per cell in mock and SNX17 knockdown cells with or without LRP1 uptake. Two-tailed t tests were performed to derive p values. n.s., not significant. F, immunoblot showing reduced SNX17 expression in CRISPR/Cas9 gene-edited NIH3T3 EHD1-GFP cells and densitometric quantification of SNX17 protein levels in cells subjected to SNX17 siRNA treatment compared with untreated cells (mock) plotted. Error bars denote standard deviation, and p values were determined by Student's two-tailed t test. Data shown are representative of three independent experiments.