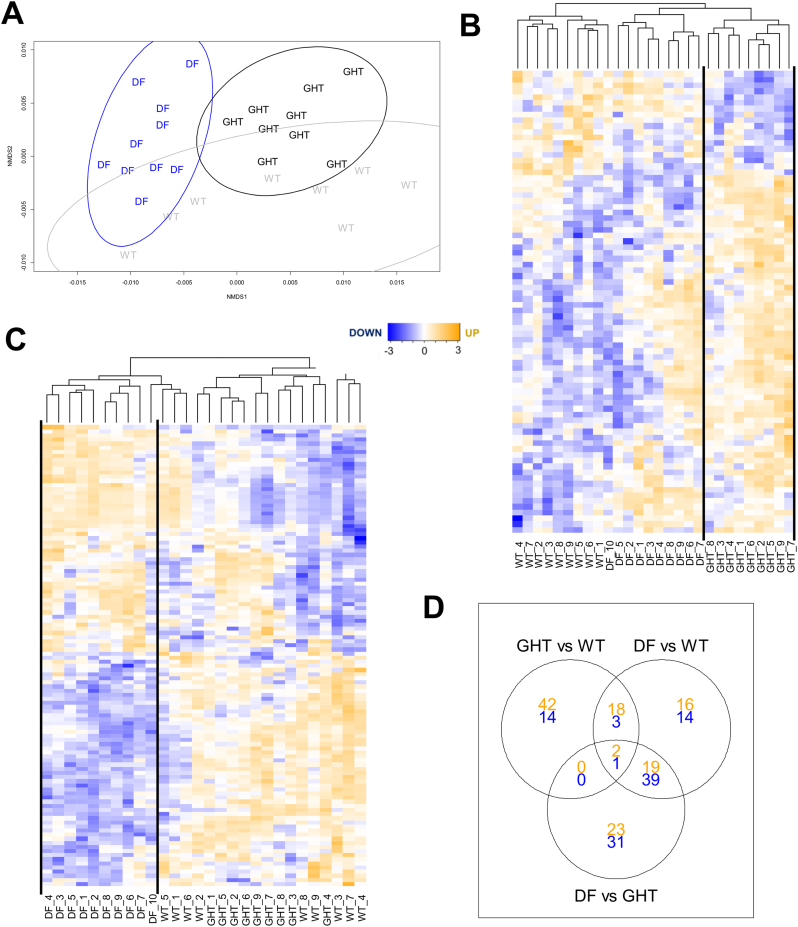
Fig. 1.
(A). Proteome-wide nonmetric multidimensional scaling analysis of wild-type (WT), growth-hormone transgenic (GHT), and domesticated fish (DF) possessing distinct growth rates. Each label represents an individual fish and ellipses represent 95% confidence intervals around strain groupings. Also shown is a hierarchical clustering and heatmap analysis demonstrating global differences in proteins showing significantly different abundances comparing (B) GHT and WT to (C) DF and WT. In each heatmap, all fish individuals from the three different strains are included for comparison. Rows represent normalized Z-scores of label-free quantification (LFQ) values from MaxQuant. Separate heatmaps for the three pairwise strain comparisons (i.e. A and B above, as well as GHT vs. DF) including protein identifications are provided in Figs. S3–5. (D) Venn diagram showing the number of significantly different protein abundances common to each pairwise strain comparison (orange: upregulated; blue: downregulated). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)