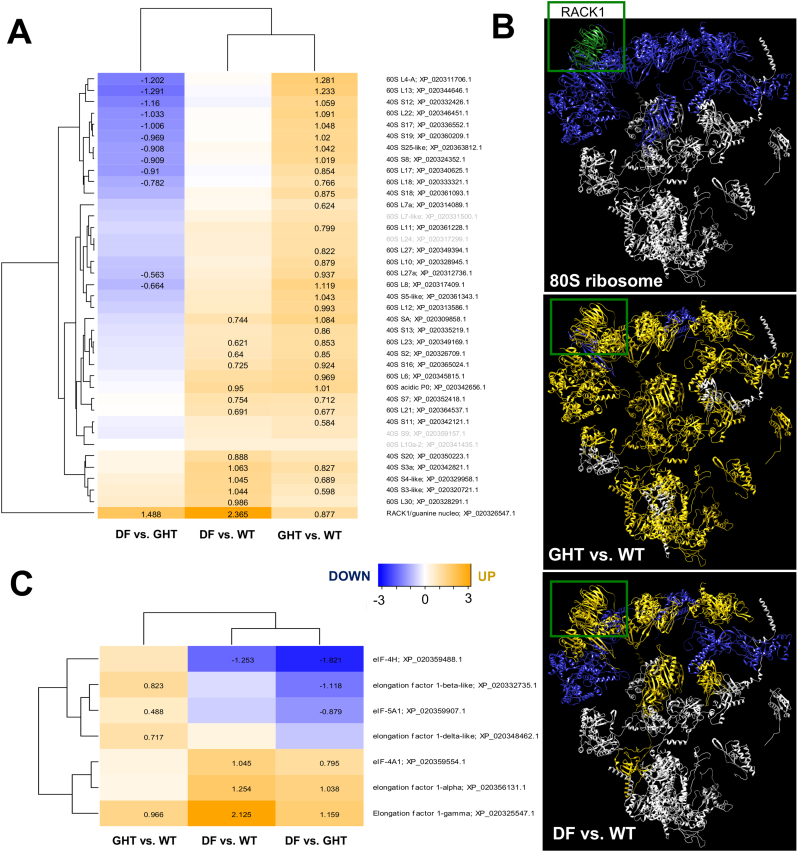
Fig. 2.
Differences in the abundance of proteins from systems driving protein synthesis in GHT and DF compared to WT. (A) Hierarchical clustering of all identified ribosomal proteins. Proteins with titles in black showed significant differential abundance across the salmon strains. Log2 fold-changes are given for proteins with significant differential abundance between the shown pairwise strain comparisons. Accession numbers and protein names are from the O. kisutch and S. salar NCBI RefSeq databases, respectively. (B) The functional relevance of changes in ribosomal protein abundance was explored by mapping the data onto a 3D structure of the human 80S ribosome [44], where any ribosomal proteins lacking orthologous salmon proteins in our dataset were removed, along with ribosomal RNA. The top panel shows all proteins orthologous to proteins in the human 80S structure, with 40S (small subunit) and 60S (large subunit) proteins shaded blue and white respectively. RACK1 is shown in green. In the lower panels, proteins showing upregulation in GHT or DF compared to WT are shaded gold. (C) Hierarchical clustering of eukaryotic translation elongation and initiation factors with details as for part B. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)