Figure 1. AD-associated genes whose products can reportedly localize to mitochondria or influence mitochondrial function.
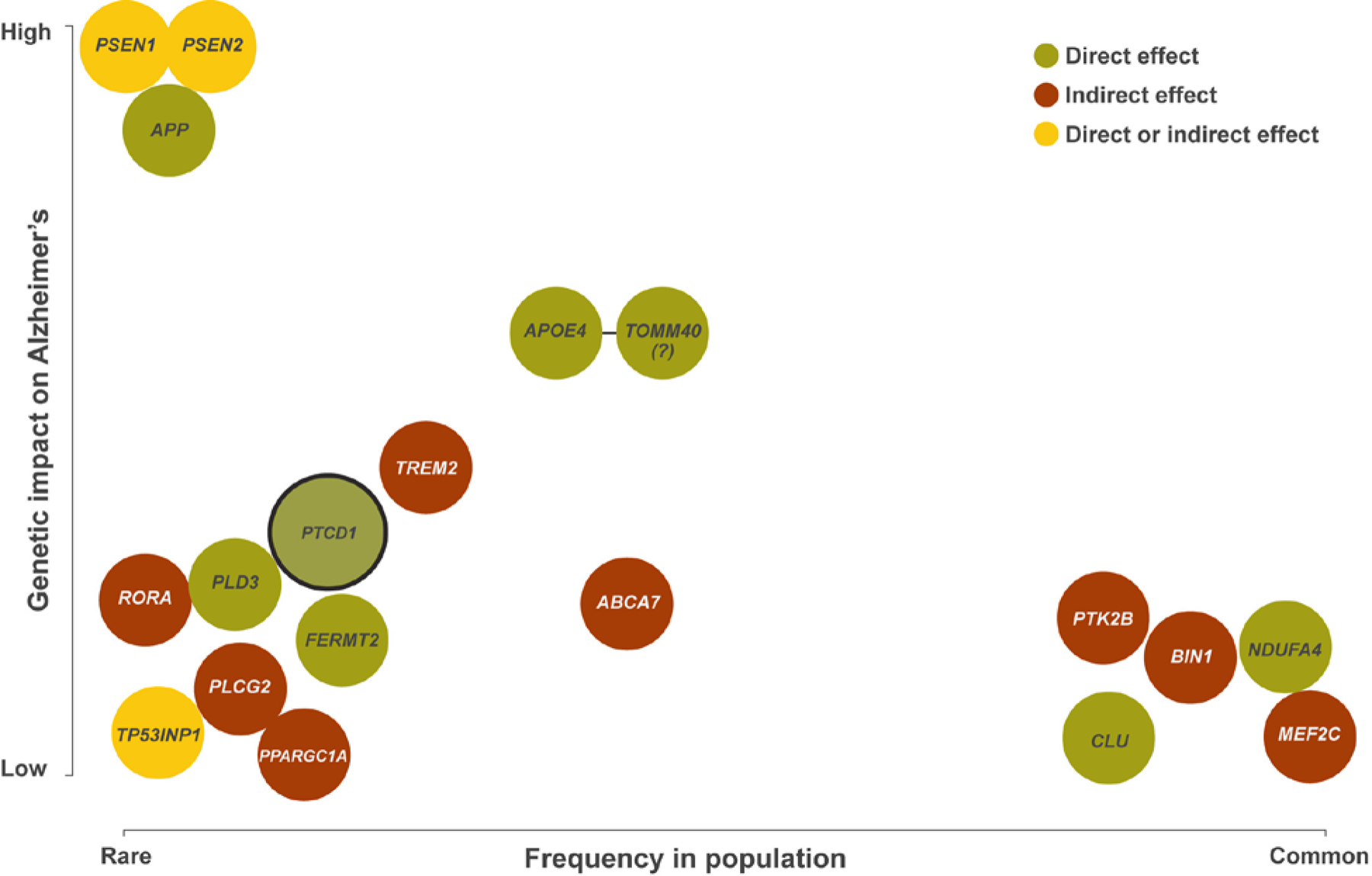
This figure includes genes said to associate with AD, usually through mechanisms unrelated to mitochondria (PTCD1 is the obvious exception), but which could plausibly affect mitochondria. The figure indicates how relatively common the variant form of the gene is in the general population (x-axis), and the relative magnitude of the impact the variant form of the gene appears to have on disease risk (y-axis). Whereas locus detection is unbiased, assignment of the responsible gene is not (in one recent AD GWAS 18 of 24 loci had a gene encoding a mitochondrial-localized protein within a 1 megabase window) [2]. Similarly, biases can be involved in the proposed biologic explanations for why a specific gene or its product influences AD risk. Current studies indicate, or are consistent with the possibility, that multiple AD-associated genes directly or indirectly affect mitochondrial function. The genes shown here were detected through a variety of approaches including linkage, GWAS, WES, and gene-based analysis. A green background indicates a potential direct effect on mitochondria, a red background indicates a potential indirect effect, and a yellow background indicates potential direct and/or indirect effects. TOMM40 is indicated with a question mark because TOMM40 is in linkage disequilibrium with APOE, and it is unclear whether TOMM40 variants independently impact AD risk.
