Abstract
Infectious bronchitis viruses (IBVs) are group III coronaviruses that infect poultry worldwide. Genetic variations, including whole-gene deletions, are key to IBV evolution. Australian subgroup 2 IBVs contain sequence insertions and multiple gene deletions that have resulted in a substantial genomic divergence from international IBVs. The genomic variations present in Australian IBVs were investigated and compared to those of another group III coronavirus, turkey coronavirus (TCoV). Open reading frames (ORFs) found throughout the genome of Australian IBVs were analogous in sequence and position to TCoV ORFs, except for ORF 4b, which appeared to be translocated to a different position in the subgroup 2 strains. Subgroup 2 strains were previously reported to lack genes 3a, 3b and 5a, with some also lacking 5b. Of these, however, genes 3b and 5b were found to be present but contained various mutations that may affect transcription. In this study, it was found that subgroup 2 IBVs have undergone a more substantial genomic rearrangements than previously thought.
Keywords: Initiation Codon, Infectious Bronchitis Virus, Predicted Amino Acid Sequence, Nucleotide Sequence Alignment, Infectious Bronchitis Virus Strain
Introduction
Infectious bronchitis virus (IBV) is a group III virus belonging to the genus Gammacoronavirus, family Coronaviridae, order Nidovirales. Numerous strains and serotypes of IBV have been, and continue to be, discovered in poultry flocks worldwide [1–6]. This genetic and antigenic diversity demonstrates the capacity of IBV to mutate extensively while maintaining the integrity of its large, single-stranded RNA genome. Mutations and recombination at various locations throughout the IBV genome are believed to modify tissue tropism and antigenicity [7–12] and are the main mechanisms responsible for the evolution of new coronaviruses that infect the same, or new, host [13, 14].
Historically, Australian IBV strains were typed into 9 distinct groups based on cross-neutralisation assays [15]. Australian IBV strains detected prior to the 1980’s, including Australian vaccine strains, were termed ‘classical’ IBV strains. These strains share common features, including tropism for both kidney and respiratory tract tissue, along with limited antigenic and S1 amino acid sequence variation (between 1 and 25%) (the IBV S1 gene forms the epitope on the surface of a mature virion particle and is transcribed as part of gene S). Between 1988 and 1994, Ignjatovic et al. [16] discovered a group of IBVs that were substantially different to ‘classical’ IBVs. These IBVs showed tropism for the respiratory tract only, no antigenic similarity with ‘classical’ strains and S1 sequence variation of greater that 44% [16, 17]. These new viruses were subsequently termed ‘novel’ IBV strains. In 2002, another group of antigenically different IBV strains was identified in Australia [18]. These viruses evolved through recombination between ‘classical’ and ‘novel’ strains [20]. Therefore, a new naming system was adopted for Australian IBVs to accommodate these established antigenically diverse groups while at the same time allowing for future isolation of new subgroups of IBVs. This naming system defines ‘classical’, ‘novel’ and 2002 strains as Australian IBV subgroup strains 1, 2 and 3, respectively [18]. These subgroups were characterised further by Hewson et al. [19] in 2009.
Viruses related to subgroup 1 and 3 strains continue to be detected in Australia; however, there has been no report on detection of subgroup 2 strains since the early 1990 s (Jagoda Ignjatovic, personal communication). Recently, recombination analysis of the Australian subgroup 3 strains has shown that while their S1 genes are closely related to those of subgroup 2 strains, the remainder of the genome is more closely related to that of the subgroup 1 vaccine strains [20]. Australian IBV strains that do not have a high S1 gene sequence identity to any current subgroups are classified as ‘other’ [19]. In this report, characterised Australian IBV isolates have been referred to as Australian reference strains and include strains from all subgroups, as well as ‘other’ strains.
The 3’ end of the IBV genome contains the main structural genes, spike (S), envelope (E), membrane (M) and nucleocapsid (N), along with several accessory genes, usually in the order S-3-E-M-5-N. Non-structural accessory protein genes 3 and 5 are polycistronic and encode proteins 3a, 3b, 5a and 5b [21, 22]. Gene 3 also encodes 3c, which has been identified as the structural envelope protein, E [21, 23, 24]. This typical organisation of coronavirus structural genes is also found in turkey coronaviruses (TCoVs) which are also group III coronaviruses that are genetically closely related to IBV. However, the genomic organisation of Australian subgroup 2 IBVs have been found to vary substantially from this typical gene organisation, and have been shown to be either S-X1-E-M-5b-N or S-X1-E-M-5b-N [25], where X1 represents a section of sequence that shows no homology to sequences from previously characterised IBVs. It has been shown, however, that the specific arrangement of these structural genes is unnecessary for the production of viable coronavirus particles [22, 26].
The genome sequence between the IBV and TCoV M and 5 genes varies in length, but on average is approximately 260-300 bp. When present, this region is referred to as an ‘intergenic region’ [27, 28] and has been suggested as a site for recombination in IBV [27]. Recently, the whole genome of TCoV was sequenced for the first time in two separate studies using three different strains of TCoV [29, 30], and two open reading frames (ORFs) (designated ORFs 4b and 4c) were discovered in this ‘intergenic region’ of TCoV [29]. Another ORF was also described, located between the N gene and 3’UTR, and designated ‘6b’. Both of these TCoV investigations compared the nucleotide sequence and location of the 4b, 4c and 6b ORFs (as designated in ref. [29]) with those of common international IBV strains, such as Beaudette and M41, and reported similar ORFs in analogous positions in the genomes of these IBV strains, but noted functional ORFs have not yet been reported in these regions of IBV. The presence of these ORFs has not been reported in Australian IBV strains, and only one investigation has examined differences present in the 3’ UTR of Australian strains [31].
The purpose of this investigation was to characterise the currently uncharacterised regions of Australian reference strains, namely the intergenic region and 3’UTR, but also the X1 region of subgroup 2 strains. ORFs discovered in these regions were compared to the sequences of the ORFs described in TCoV. Analysis of the ORFs present in the genome of Australian subgroup 2 strains was of particular interest, as sections of the genome corresponding to the ORFs in the TCoV intergenic region appear to be absent in this subgroup [25].
Materials and methods
Nomenclature employed by Cao et al. [29] for the TCoV ORFs was used in this study.
Analysis of the X1 genomic region of the Australian subgroup 2 genome
Sequence data from Australian subgroup 2 isolates N1/88, Q3/88, V18/91 and V6/92 were obtained from GenBank [32] (accession numbers DQ490207, DQ490212, DQ490219 and DQ490220, respectively). This represents all known Australian subgroup 2 isolates [25]. The nucleotide sequence from 150 bp upstream of the S gene termination codon to 150 bp downstream of the initiation codon of the E gene for each of the subgroup 2 reference strains was translated and scanned for ORFs using ORF Finder (www.ncbi.nlm.nih.gov). The nucleotide sequence of each of the detected ORFs was compared to sequences available publicly in the GenBank database, using the a nucleotide BLAST search (blastn / megablast) against the ‘others’ database and the predicted translated sequence was subjected to a protein-protein BLAST search (blastp) against the ‘non-redundant protein sequence’ database. Amino acid and nucleotide sequence alignments were performed using Clustal W2 [33].
The hydrophobic and hydrophilic regions of the predicted proteins encoded by the detected ORFs were compared using a hydropathicity plot generator (http://www.vivo.colostate.edu/molkit/hydropathy/) with the Kyte-Doolittle and Hopp-Woods algorithms.
Analysis of the intergenic region of Australian subgroup 2 and Australian reference strains of IBV
The nucleotide sequence from 150 bp upstream of the M gene termination codon to 150 bp downstream of the nucleocapsid gene initiation codon for each of the subgroup 2 strains, Australian vaccine strains (VicS, S and Armidale; GenBank accession numbers DQ490221, DQ490213 and DQ490205 respectively), Australian subgroup 1 strains (Q1/76, Q1/99 and V5/90; GenBank accession numbers DQ490210, DQ490211, and DQ490218 respectively) and Australian ‘other’ strains (Q1/73, N1/62, N2/75, V1/71, V2/71, V2/02 and V3/02; GenBank accession numbers DQ490209, DQ490206, DQ490208, DQ490214, DQ490216, DQ490215 and DQ490217 respectively) was extracted from each complete structural gene sequence located in GenBank. The nucleotide sequence from each of these viruses was translated, scanned for ORFs and subjected to blastn and blastp searches as described above. Sequence alignments were performed using ClustalW2 as described above.
Results
The X1 region of the subgroup 2 genome is comprised of two complete ORFs, one of which appears to have been translocated
Schematic diagrams of the gene arrangements for TCoV [29] and Australian subgroup 1 and 2 strains are presented in Fig. 1.
Fig. 1.
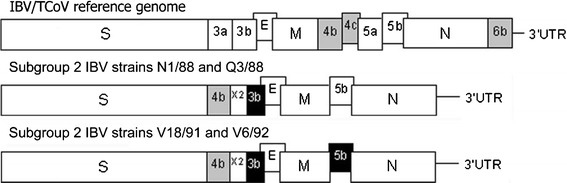
Schematic representation of the arrangement of the structural genes of IBV and TCoV. S, spike; 3a, 3b and E, gene 3; M, membrane; 4b, 4c ORFs (‘intergenic region’); 5a and 5b, gene 5; N, nucleocapsid; 6b ORF; X2, no identity with any sequence in GenBank. N1/88, Q3/88, V18/91 and V6/92 are Australian subgroup 2 IBV strains. Uncharacterised ORFs are represented by grey boxes, and genes containing mutations that may affect transcription are represented by black boxes
Blastp and ORF analyses of the translated nucleotide sequence between the S and E genes of the four subgroup 2 strains, referred to as X1 by Mardani et al. [25], identified two ORFs. The first was found 13 bp downstream of the S gene termination codon. This ORF was 264 bp in length and present in all subgroup 2 strains. The predicted amino acid sequences for these ORFs had sequence identities of 89-100% between subgroup 2 strains and identities of 36-43% with TCoV ORF 4b (GenBank accession numbers ABW75129 and ABW75143). Therefore, the location of a 4b-like ORF in the subgroup 2 IBV strains was different to the location of ORF 4b in TCoV, as it was found between the S and E genes instead of between the M and 5 genes. (see Fig. 1). Alignment of the predicted amino acid sequence encoded by this subgroup 2 4b-like ORF with the homologous sequence in 13 other Australian reference strains is presented in Fig. 2.
Fig. 2.
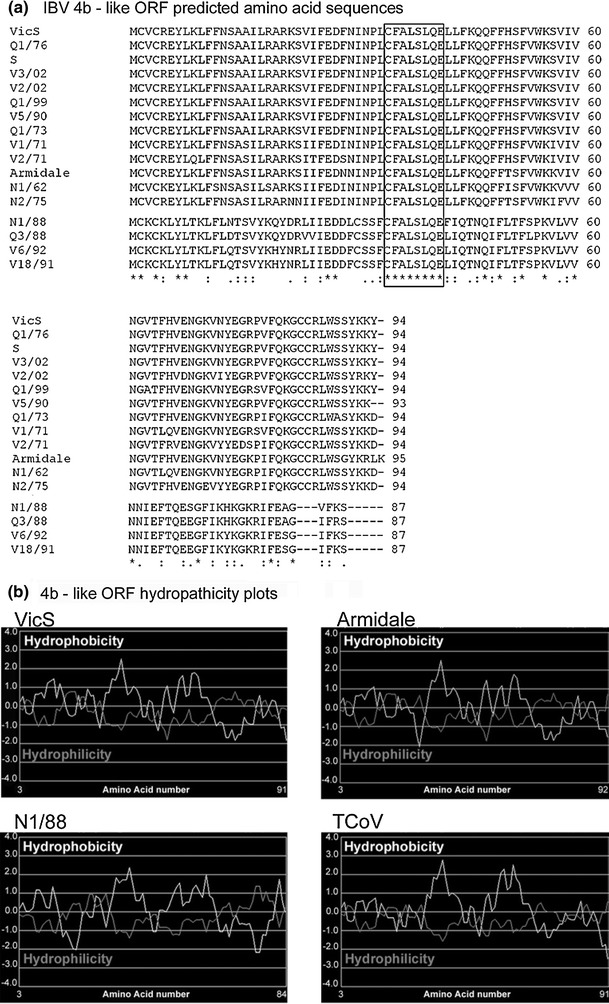
Comparison of the 4b-like ORFs in Australian IBVs. a) Alignment of the predicted amino acid sequences of the 4b-like ORFs found in Australian IBV strains. A blank line has been introduced to distinguish between subgroup 2 strains (below the line) with Australian reference IBVs (above the line). Gaps are represented by dashes (-), and identical amino acids are indicated with an asterisk (*). The protein motif “CFALSLQE”, which is conserved in all Australian IBV strains, is boxed. Conserved amino acid changes are represented with “:”, while semi-conserved amino acid changes are identified by “.”. b) Hydropathicity plots for Australian IBV vaccine strains VicS and Armidale, subgroup 2 strain N1/88 and TCoV strain ATCC. Hydrophobicity (Kyte-Doolittle scale) is represented by the white line, while hydrophilicity (Hopp-Woods scale) is represented by the grey line. The plots were generated using http://www.vivo.colostate.edu/molkit/hydropathy/
Downstream from the 4b-like ORF, but in a different frame, a region of 342 bp (321 bp for strain Q3/88, which had a 21-bp deletion) was detected. In all subgroup 2 strains the 183 bp at the 3’ end of this region was similar to, and encompassed, the entire IBV 3b gene. The alignment of the predicted amino acid sequences of the subgroup 2 3b genes with homologous sequences from Australian reference IBV strains is presented in Fig. 3. The nucleotide sequence identities generated for this gene were 91-100% between the subgroup 2 strains and 31-38% with the 3b genes of the subgroup 1 Australian strains. The first codon in the 3b gene of the subgroup 2 strains was GTA, which encodes valine, instead of an ATG initiation codon (Fig. 3).
Fig. 3.

Alignment of the predicted amino acid sequences of the subgroup 2 3b genes with those of vaccine strains VicS and Armidale. a) Alignment of the predicted amino acid sequences of gene 3b of Australian subgroup 2 strains N1/88, Q3/88 V18/91 and V6/92 with Australian vaccine IBV strains VicS and Armidale. Identical amino acids and bases are indicated by an asterisk (*). Conserved amino acid changes are represented by “:”, while semi-conserved amino acid changes are identified by “.”. b) Nucleotide sequence alignment of the first three codons of gene 3b. The initiation codon is boxed to show the altered initiation codon in the subgroup 2 strains
The remaining 159 bp (138 bp for Q3/88) of the 339-bp region, downstream of the 4b-like ORF and upstream of gene 3b, encoded a predicted peptide that extended from an asparagine (codon AAU) to a glutamine (codon CAA). Analysis of both the nucleotide and predicted amino acid sequence identified no significant identities with any sequence published to date. This region was therefore designated the subgroup 2 X2 region.
The 5b gene is present but truncated in the subgroup 2 strains V6/91 and V18/92
The region between the M and N genes for subgroup 2 strains was determined to be 219 bp for N1/88 and Q3/88, and 212 bp and 179 bp for V18/91 and V6/92, respectively. A previous study analysing this region in subgroup 2 viruses reported that N1/88 and Q3/88 both contained a 5b gene only in this region, while V6/92 and V18/91 strains lacked any ORF in this region [25]. The 5b gene in N1/88 and Q3/88 is 246 bp, beginning 37 bp downstream of the M gene termination codon and ending 61 bp downstream of the N gene initiation codon.
In this study, analysis of the genomic region between the M and N genes in strain V18/91 resulted in detection of an ORF of 105 bp, with the initiation codon 30 bp downstream of the M gene termination codon. This ORF was predicted to encode 34 amino acids, and Blastp analysis showed that the first 22 amino acids were identical to the first 22 amino acids of the N1/88 5b gene product. In another frame of this region of V18/91, there were two additional ORFs of 150 bp and 102 bp, separated by a single termination signal due to a cytosine-to-thymidine substitution. Analysis of the predicted amino acid sequence for the 150-bp ORF revealed that the last 45 amino acids had 75% sequence identity to amino acids 22–66 of both the N1/88 and Q3/88 5b genes. Analysis of the predicted amino acid sequence predicted for the 102-bp ORF revealed that the first 14 amino acids aligned with the last 14 amino acids encoded by the N1/88 5b gene (amino acids 68-81). A thymidine-to-cytosine substitution eliminated the termination codon in V18/91, resulting in an 18-amino-acid extension upstream of the 5b gene termination signal of N1/88 and Q3/88. Blastp analysis showed that these 18 amino acids had no significant similarity to any sequences in GenBank.
Therefore, V18/91 was shown to contain a 5b gene, albeit a truncated one. The nucleotide sequence alignment of the V18/91 5b gene with N1/88 and Q3/88 revealed a single base insertion in an adenosine tandem repeat downstream of the initiation codon in the first ORF. This insertion resulted in a frameshift and resultant termination 36 bp downstream of the tandem repeat.
The genomic sequence between the M and N genes in strain V6/92 was found to contain two ORFs of 72 bp and 267 bp, in different frames. The 72-bp ORF had an initiation codon 30 bp downstream of the M gene termination signal. The first 12 amino acids of the predicted peptide for this ORF were identical to the first 12 amino acids of the N1/88 5b gene. Analysis of the predicted product of the 267-bp ORF of V6/92 showed that the central 58 amino acid residues had sequence identities of 77% and 74% with the amino terminus of the predicted 5b protein of the N1/88 and Q3/88 strain, respectively. As with V18/91, a thymidine-to-cytosine substitution eliminated the termination codon, resulting in an 18-amino-acid extension. This 18-amino-acid extension in V6/92 was 100% identical to the 18-amino-acid extension found in V18/91.
The nucleotide sequence alignment of the V6/92 5b gene with N1/88 and Q3/88 revealed a 34-bp deletion 36 bp after the initiation methionine. It appears that either this deletion was incomplete, leaving a single guanosine (G37), or a guanosine was inserted after a 35-bp deletion, but either way, it resulted in a truncated V6/92 5b gene.
Additional ORFs 4b, 4c and 6b are located in the structural gene region of Australian IBV strains
The genomic region between the M and 5a genes of Australian reference IBV strains (excluding subgroup 2 strains) ranged from 359 to 363 bp. Sequence analysis of this region predicted the presence of at least two ORFs. In all strains, the initiation codon for the first ORF was located immediately after the M gene termination codon, in the same frame. This first ORF was predicted to code for 94 amino acids in all strains, except for V5/90 and Armidale, in which this ORF coded for 93 and 95 amino acids, respectively. Amino acid identities between Australian strains in this region ranged from 82 to 100%. Sequence identities were found with TCoV ORF 4b only and ranged from 76 to 87%. This same region had amino acid sequence identities of 33-39% with the 87-amino-acid-long 4b-like ORF detected in the subgroup 2 strains, which was located immediately after the S gene. Therefore, the location of the 4b-like ORF in all Australian strains, except the subgroup 2 strains, was the same as that of the TCoV 4b gene (immediately downstream of the 5b gene). An alignment of the predicted amino acid sequences of the 4b-like ORF is shown in Fig. 2. The genomic sequence of the IBV reference strain, Beaudette, contains a truncated 4b-like ORF of 50 amino acids (results not shown).
An alignment of the predicted amino acid sequences for the 4b-like ORFs of all strains analysed in this study (including the subgroup 2 strains) showed a conserved (100% identity) sequence of 8 residues, CFALSLQE (Fig. 2), located 35 amino acids downstream of the initiation methionine. When this alignment was extended to include the predicted amino acid sequence of the TCoV 4b ORF, the sequence was CFILSFQE, which was identical to the region found in IBV save for two amino acid changes – one conserved, one non-conserved (not shown).
Hydropathicity plots of the predicted proteins encoded by the 4b-like ORFs showed two hydrophobic regions in similar positions in all IBV strains analysed. Hydropathicity plots were very similar for different IBV strains, including the subgroup 2 strains, and similar for the predicted product of the TCoV ORF 4b (Fig. 2).
An additional ORF of 171 bp was detected in the region between the M and 5 genes of all Australian IBV strains, excluding subgroup 2 strains. This ORF was located downstream of the 4b-like ORF and was 171 bp in length in all strains except V5/90 and Armidale where this ORF was 81 bp and 78 bp in length, respectively. The full-length 171-bp ORF was predicted to code for a 56-amino-acid-long peptide, with the initiation codon located 80 bp before the termination codon of the 4b-like ORF. The termination codon for this ORF was 17 bp after the start of the 5a gene. Analysis of the predicted amino acid sequence showed high identity only with the complete sequence of TCoV ORF 4c [29]. Amino acid sequence identities were 78-100% between the Australian IBVs and 56-76% with TCoV ORF 4c (GenBank accession numbers ABW75130 and ABW75144).
A third ORF, which was predicted to code for 72–74 amino acids in strains Armidale, V2/02, V3/02, Q1/73, N1/62, V1/71 and V2/71, was also detected downstream of the nucleocapsid gene termination codon. All other Australian strains analysed in this study had truncated versions of this ORF, of varying lengths. Analysis of the predicted coding sequence showed sequence similarity with the entire TCoV ORF 6b [29]. Amino acid sequence identities of 80-87% were found between the TCoV ORF 6b and those of the Australian strains. Between Australian strains, the level of sequence identity was 82-100%. Blastn analysis of the 6b-like ORF nucleotide sequences detected in Australian IBV strains revealed identities with the region downstream of the nucleocapsid gene in several international strains (results not shown).
Discussion
In this investigation, ORFs were detected in Australian IBV strains that have a high level of sequence identity to ORFs recently identified in the structural gene region of the TCoV genome [29, 30]. Further, it was also found that the gene organisation in Australian subgroup 2 IBV strains is even more diverse than previously reported [25].
The ORF designated 4b in TCoV was present in all Australian IBV strains analysed in this study, with high sequence similarities. The location of this ORF was between genes M and 5 for TCoV [29, 30] and all Australian reference strains, with the exception of the subgroup 2 strains. The 4b-like ORF in subgroup 2 strains was located immediately downstream of, and in the same frame as, the S gene. This indicated that the previously reported subgroup 2 X1 region [25] was partially comprised of a 4b-like ORF.
The unique position of the 4b-like ORF in subgroup 2 viruses provides evidence that translocation may have occurred within the IBV genome. This would be the first description of a coronavirus using translocation as a mutation mechanism in addition to the widely reported mechanisms of whole-gene deletions, point mutations and recombination [7, 8, 11, 27, 28, 34–39]. It is interesting to note that the subgroup 2 genome organisation is more similar to the genomic organisation of mammalian coronaviruses [40] than the genome organisation of IBV and TCoV. Therefore, the results of this study may also suggest that Australian subgroup 2 IBVs perhaps did not emerge from group III coronaviruses. The findings of this investigation, therefore, have implications not only for IBV research but also for research on coronaviruses in general.
It is important to note that the subgroup 2 strains have completely retained the 4b-like ORF coding region, along with their ability to infect and cause disease in chickens, despite the loss of some non-structural accessory protein genes and extensive genome rearrangements and deletions [16]. The identity scores between the 4b ORF of TCoV and the 4b-like ORFs in Australian IBV strains (including subgroup 2 strains) suggested a genomic similarity between these ORFs, and these results, coupled with the similarity between the hydropathicity plots of their respective predicted proteins, suggest that their products would fold similarly and therefore retain similar properties. The presence of this complete ORF in many international and most Australian IBV strains, as well as the presence of a highly conserved region within this ORF, suggests an important functional role in IBVs. Although, to date, no confirmed functional ORF has been found between the M and 5a genes in IBV or TCoV, the findings of this study, combined with the conclusions drawn by Cao et al. [29] and Gomaa et al. [30], indicate that future investigations aimed at defining the function of the 4b-like ORF are warranted.
Open reading frames homologous to TCoV ORFs 4c and 6b were detected in most of the Australian IBV strains analysed in this study. In some cases, these ORFs were present but truncated, although all were located in analogous positions to ORFs 4c and 6b in TCoV [29]. Neither of these ORFs was present in subgroup 2 viruses.
Despite a previous report concluding that the subgroup 2 strains had no 3b gene present in their genome [25], this investigation showed that this was not the case. All subgroup 2 strains were found to contain a homologue of gene 3b, but it appeared that the initiation codon had been inverted. In subgroup 2 viruses, the region denoted X2 was positioned upstream from gene 3b and downstream of the 4b-like ORF. The X2 region may be either a remnant from the translocation event or the remainder of the 3a gene, which is found upstream of the 3b gene in other IBVs but is absent in all subgroup 2 strains. This investigation has shown that the X1 region in subgroup 2 strains is comprised of two ORFs separated by an unknown region, in the order 4b – X2 – 3b.
In further contrast to the previous report [25], subgroup 2 strains V6/92 and V18/91 were found to contain a 5b gene; however, nucleotide insertions and/or deletions had resulted in premature termination of this gene in both of these strains. Further work will be needed to determine if the 3b genes and the truncated 5b genes in subgroup 2 viruses are functional.
Previous studies have sought to identify the role of the IBV non-structural accessory proteins (3a, 3b, 5a and 5b) by genetically modifying the Beaudette reference strain of IBV, but these studies resulted in the generation of an attenuated non-pathogenic, Beaudette strain of IBV [21, 22, 24, 41]. Australian subgroup 2 strains are naturally occurring viruses and therefore provide a useful alternative model for elucidating the role of these genes in the natural host.
Acknowledgments
The primary author of this investigation was supported by an Australian Postgraduate Award and a scholarship from the Australian Poultry Cooperative Research Centre. The Authors would like to thank the Asia Pacific Centre for Animal Health for their support.
References
- 1.Cavanagh D, Davis PJ, Cook JKA, et al. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21:33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- 2.Cavanagh D, Davis PJ, Mockett APA. Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes. Virus Res. 1988;11:141–150. doi: 10.1016/0168-1702(88)90039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolz R, Pujols J, Ordóñez G, et al. Antigenic and molecular characterization of isolates of the Italy 02 infectious bronchitis virus genotype. Avian Pathol. 2006;35:77–85. doi: 10.1080/03079450600597295. [DOI] [PubMed] [Google Scholar]
- 4.Farsang A, Ros C, Renström LHM, et al. Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement of a vaccine strain. Avian Pathol. 2002;31:229–236. doi: 10.1080/03079450220136530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gelb J, Jr, Wolff JB, Moran CA. Variant Serotypes of Infectious Bronchitis Virus Isolated from Commercial Layer and Broiler Chickens. Avian Dis. 1991;35:82–87. doi: 10.2307/1591298. [DOI] [PubMed] [Google Scholar]
- 6.Wang CH, Huang YC. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch Virol. 2000;145:291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ammayappan A, Upadhyay C, Gelb J, et al. Complete genomic sequence analysis of infectious bronchitis virus Ark DPI strain and its evolution by recombination. Virol J. 2008;5:157–163. doi: 10.1186/1743-422X-5-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochkov YA, Tosi G, Massi P, et al. Phylogenetic analysis of partial S1 and N gene sequences of infectious bronchitis virus isolates from Italy revealed genetic diversity and recombination. Virus Genes. 2007;35:65–71. doi: 10.1007/s11262-006-0037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen HW, Huang YP, Wang CH. Identification of Taiwan and China-like recombinant avian infectious bronchitis viruses in Taiwan. Virus Res. 2009;140:121–129. doi: 10.1016/j.virusres.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolz R, Pujols J, Ordóñez G, et al. Molecular epidemiology and evolution of avian infectious bronchitis virus in Spain over a fourteen-year period. Virology. 2008;374:50–59. doi: 10.1016/j.virol.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, Zhang Q, Chen J, et al. Identification of the avian infectious bronchitis coronaviruses with mutations in gene 3. Gene. 2008;412:12–25. doi: 10.1016/j.gene.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Junker D, Collisson EW. Evidence of natural recombination within the S1 genes of infectious bronchitis virus. Virology. 1993;192:710–716. doi: 10.1006/viro.1993.1093. [DOI] [PubMed] [Google Scholar]
- 13.Vijgen L, Keyaerts E, Van Ranst M. Molecular evolution of group 2 coronaviruses. In: Thiel V, editor. Coronaviruses: Molecular and Cellular Biology. Norfolk: Caister Academic Press; 2007. pp. 143–158. [Google Scholar]
- 14.Snijder EJ, Bredenbeek PJ, Dobbe JC, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wadey CN, Faragher JT. Australian infectious bronchitis viruses: identification of nine subtypes by a neutralisation test. Res Vet Sci. 1981;30:70–74. doi: 10.1016/S0034-5288(18)32611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ignjatovic J, Sapats SI, Ashton F. A long-term study of Australian infectious bronchitis viruses indicates a major antigenic change. Avian Pathol. 1997;26:535–553. doi: 10.1080/03079459708419233. [DOI] [PubMed] [Google Scholar]
- 17.Sapats SI, Ashton F, Wright PJ, et al. Sequence analysis of the S1 glycoprotein of infectious bronchitis viruses: identification of a novel genotypic group in Australia. J Gen Virol. 1996;77:413–418. doi: 10.1099/0022-1317-77-3-413. [DOI] [PubMed] [Google Scholar]
- 18.Ignjatovic J, Gould G, Sapats S. Isolation of a variant infectious bronchitis virus in Australia that further illustrates diversity among emerging strains. Arch Virol. 2006;151:1567–1585. doi: 10.1007/s00705-006-0726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewson K, Noormohammadi A, Devlin J, et al. Rapid detection and non-subjective characterisation of infectious bronchitis virus isolates using high-resolution melt curve analysis and a mathematical model. Arch Virol. 2009;154:649–660. doi: 10.1007/s00705-009-0357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mardani K, Noormohammadi A, Ignjatovic J et al (2010) Naturally occurring recombination between distant strains of infectious bronchitis virus. Arch Virol [DOI] [PMC free article] [PubMed]
- 21.Britton P, Casais R, Hodgson T et al (2006) Genes 3 and 5 of Infectious Bronchitis Virus are Accessory Protein Genes. In: Perlman S and Holmes K V (ed) The Nidoviruses, Springer US363-368 [DOI] [PMC free article] [PubMed]
- 22.Hodgson T, Britton P, Cavanagh D. Neither the RNA nor the Proteins of Open Reading Frames 3a and 3b of the Coronavirus Infectious Bronchitis Virus Are Essential for Replication. J Virol. 2006;80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boursnell MEG, Brown TDK, Foulds IJ, et al. Completion of the Sequence of the Genome of the Coronavirus Avian Infectious Bronchitis Virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 24.Cavanagh D, Casais R, Armesto M, et al. Manipulation of the infectious bronchitis coronavirus genome for vaccine development and analysis of the accessory proteins. Vaccine. 2007;25:5558–5562. doi: 10.1016/j.vaccine.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mardani K, Noormohammadi AH, Hooper P, et al. Infectious bronchitis viruses with a novel genomic organization. J Virol. 2008;82:2013–2024. doi: 10.1128/JVI.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Haan CAM, Volders H, Koetzner CA, et al. Coronaviruses Maintain Viability despite Dramatic Rearrangements of the Strictly Conserved Genome Organization. J Virol. 2002;76:12491–12502. doi: 10.1128/JVI.76.24.12491-12502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks JE, Rainer AC, Parr RL, et al. Comparisons of envelope through 5B sequences of infectious bronchitis coronaviruses indicates recombination occurs in the envelope and membrane genes. Virus Res. 2004;100:191–198. doi: 10.1016/j.virusres.2003.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavanagh D, Davis PJ. Evolution of Avian Coronavirus IBV: Sequence of the Matrix Glycoprotein Gene and Intergenic Region of Several Serotypes. J Gen Virol. 1988;69:621–629. doi: 10.1099/0022-1317-69-3-621. [DOI] [PubMed] [Google Scholar]
- 29.Cao J, Wu CC, Lin TL. Complete nucleotide sequence of polyprotein gene 1 and genome organization of turkey coronavirus. Virus Res. 2008;136:43–49. doi: 10.1016/j.virusres.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomaa MH, Barta JR, Ojkic D, et al. Complete genomic sequence of turkey coronavirus. Virus Res. 2008;135:237–246. doi: 10.1016/j.virusres.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sapats SI, Ashton F, Wright PJ, et al. Novel Variation in the N Protein of Avian Infectious Bronchitis Virus. Virology. 1996;226:412–417. doi: 10.1006/viro.1996.0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson DA, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 34.Davidson I, Silva R. Creation of diversity in the animal virus world by inter-species and intra-species recombinations: lessons learned from poultry viruses. Virus Genes. 2008;36:1–9. doi: 10.1007/s11262-007-0165-1. [DOI] [PubMed] [Google Scholar]
- 35.Herrewegh AAPM, Smeenk I, Horzinek MC, et al. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee C-W, Jackwood M. Spike Gene Analysis of the DE072 Strain of Infectious Bronchitis Virus: Origin and Evolution. Virus Genes. 2001;22:85–91. doi: 10.1023/A:1008138520451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pantin-Jackwood M, Spackman E, Woolcock P. Phylogenetic Analysis of Turkey Astroviruses Reveals Evidence of Recombination. Virus Genes. 2006;32:187–192. doi: 10.1007/s11262-005-6875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyrc K, Dijkman R, Deng L, et al. Mosaic Structure of Human Coronavirus NL63, One Thousand Years of Evolution. J Mol Biol. 2006;364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stavrinides J, Guttman DS. Mosaic Evolution of the Severe Acute Respiratory Syndrome Coronavirus. J Virol. 2004;78:76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasternak AO, Spaan WJM, Snijder EJ. Nidovirus transcription: how to make sense…? J Gen Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 41.Casais R, Davies M, Cavanagh D, et al. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J Virol. 2005;79:8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


