Fig. 2.
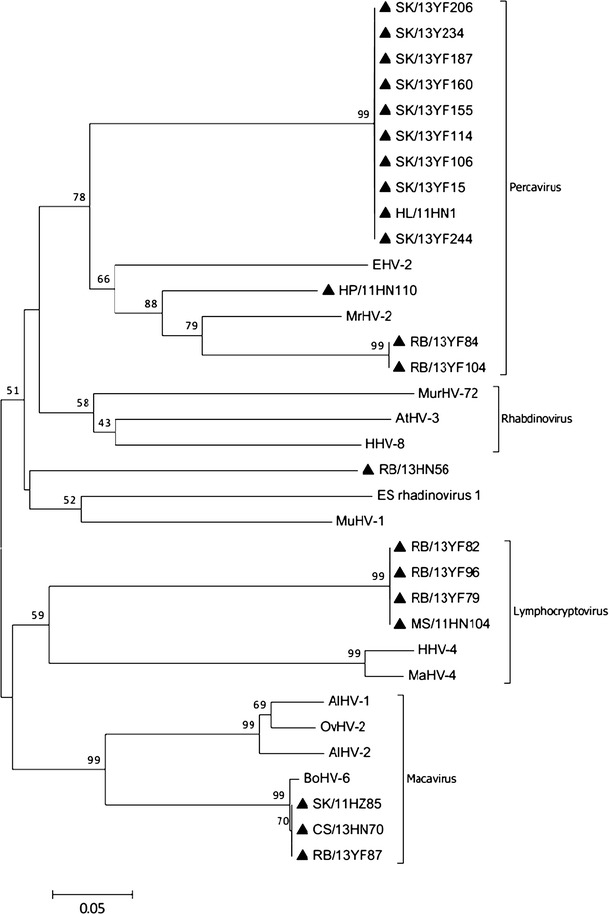
Phylogenetic analysis based on a 151-amino-acid portion of the deduced amino acid sequence of glycoprotein B (gB) of herpesviruses from bats in southern China. The tree was generated by using the neighbor-joining method with the p-distance model. A bootstrap test was replicated 1000 times. Numbers above the branches indicate NJ bootstrap values. Bold triangles indicate herpesviruses detected in the present study. CS, Cynopterus sphinx; HL, Hipposideros larvatus; HP, Hipposideros pomona; MR, Myotis ricketti; MS, Miniopterus schreibersi; RB, Rhinolophus blythi; SK, Scotophilus kuhlii; YF, Yunfu; HN, Hainan province; HZ, Huizhou
