Abstract
On the basis of partial sequencing of the infectious bronchitis virus (IBV) S1 gene, this study investigated the molecular diversity of the virus in two life periods of a batch of breeding hens at the field level. The chicks were vaccinated against IBV on the second day of life with the vaccine Ma5, but at the age of 18 days, they exhibited clinical signs and macroscopic lesions compatible with avian infectious bronchitis (IB). In the clinical disease stage, the Ma5 vaccine strain was detected in the trachea, lungs, and small intestine of the chicks, while IBV variants were detected in the bursa of Fabricius and kidneys. Subsequently, new samples were collected from the same batch at the end of the production cycle. In this phase, the Ma5 vaccine strain was detected in the kidneys, small intestine, and oviduct of the hens. However, a previously unidentified IBV variant was found in the cecal tonsils. Additionally, a fragment of viral RNA with that was completely identical to the corresponding region of the Ma5 vaccine was detected in the allantoic fluid of viable embryos from the hens under study after 18 days of incubation. These findings suggest that, in addition to the Ma5 vaccine, other strains of IBV variants can coexist, seeming to establish a chronic infection in the chickens, and that they can potentially be transmitted vertically. These results may assist in immunoprophylaxis control programs against IBV.
Electronic supplementary material
The online version of this article (doi:10.1007/s00705-016-3030-5) contains supplementary material, which is available to authorized users.
Keywords: Vertical Transmission, Infectious Bronchitis Virus, Infectious Bronchitis Virus Strain, Cecal Tonsil, Infectious Bronchitis Virus Infection
Introduction
Avian infectious bronchitis (IB) is a disease that affects fowl of all ages. It is caused by infectious bronchitis virus (IBV), a member of the family Coronaviridae [7, 22].
The genetic instability of IBV has been observed through the emergence of antigenic and genetic variants over the course of its evolution, and the alterations are mainly associated with the glycoprotein S1, which has important biological properties, including definition of cell tropism and determination of the serotype, and it is responsible for the induction of neutralizing antibodies [17].
In addition to replicating in tissues and causing prominent lesions in the respiratory tract, the genetic variability of IBV may have favored differences in the tropism and pathogenicity of many other types of epithelial cells, such as those of the kidneys [14, 36], oviduct and testes [37], and gastrointestinal tract [6], and it is associated with lesions on the pectoral musculature [2, 19]. Therefore, the importance of IB lies in the economic losses, as the infection often causes respiratory disease in broiler chickens, decreasing weight gain, increasing mortality and the rejection of slaughter carcasses, and increasing the susceptibility of the chickens to opportunistic bacterial infections [32]. In laying and breeding hens, the infection has been associated with a decrease in production, a worse internal and external quality of the eggs, and a reduction in the hatchability rate [41].
The difficulty in controlling IB is a result of the multiple unpredictable IBV variants that emerge or are introduced on poultry farms. Due to the emergence of new genotypes, pathotypes, and serotypes, IB has not yet been satisfactorily controlled and stands out as one of the most important health problems related to the reduction in the productive capacity of chickens and consequent economic losses in the poultry industry [20–22].
A characteristic of RNA viruses is their ability to adapt quickly to the host through genetic changes during their replication. As such, coronaviruses, including IBV, have been proven to exist as a mixture of genetically distinct strains, which results in a heterogeneous population of viruses transcribed from a single progenitor genome [22]. The mechanisms that regulate the evolution of IBV include mutation, recombination, and selection in the host [34, 45]. Additionally, studies have shown that the evolutionary mechanisms for the emergence of viral variants are also present in commercial live vaccines against IBV [16, 34, 39, 45].
In this study, we amplified and sequenced fragments of the IBV S1 gene taken from tissue samples from different chicken organs naturally affected by the virus. These sequences were compared with those of other IBV strains available in the GenBank database and analyzed with respect to their polymorphisms. Our objectives were (1) to assess the presence and distribution of IBV strains in different organs of chicks and hens with the clinical disease and (2) to assess the presence of viral RNA in the reproductive system and describe the evidence of vertical transmission in a natural infection.
Materials and methods
Sample collection
At the start of 2013, 18-day-old chicks belonging to a single batch of 36,000 breeding hens, located in the state of Minas Gerais, Brazil, presented clinical signs such as sneezing, polyuria, and 12 % mortality. The chicks were vaccinated by spray at 2 days of age with IBV Nobilis® IB Ma5 (MSD, Animal health). The Ma5 vaccine is produced from of a biological clone a nonpathogenic IBV strain, which is replicated in the tissues of SPF chicken embryos and marketed as a live vaccine. Samples of the trachea, lungs, kidneys, small intestine, and bursa of Fabricius of 10 chicks were obtained in the form of a pool of each organ from the batch.
At the end of the production cycle of this batch (52 weeks – 364 days of age), five hens were sacrificed to collect their kidneys, small intestine, cecal tonsils, and oviducts in the form of a pool to form a set for each organ. In addition, 10 embryonated eggs of this batch (18 days of incubation) were chilled to 4 °C for 24 h, and the chorioallantoic fluid was collected from each embryo to form a pool for the detection and identification of IBV. The samples were collected following the criteria and results established by Cook [12], Lucio and Fabricant [31], and Naqi et al. [38].
RNA extraction
In order to obtain viral RNA from the organs, a small portion of each tissue was collected. RNA extraction was performed using TRIzol® Reagent (Invitrogen™) according to the recommendations of the manufacturer. The RNA was suspended in 50 µl of UltraPure™ DEPC-treated water (Invitrogen™), analyzed and quantified by the fluorescence method using a NanoDrop Lite spectrophotometer (Thermo Scientific), and immediately used in the reverse transcription reaction.
cDNA synthesis
The IBV RNA was retrotranscribed to cDNA in a Veriti® thermal cycler (Applied Biosystems). The final volume of the reaction mixture was 20 μL, containing 700 ng of total RNA, 50 µM of the oligonucleotide S1OLIGO3’ [27], and the enzyme SuperScript® III Reverse Transcriptase (Invitrogen™), following the manufacturer’s recommendations.
Nested RT-PCR
The amplification was performed in two consecutive reactions in a Veriti® thermal cycler (Applied Biosystems). In the first reaction, the RT-PCR was performed with 25 µL of Go Taq® Green Master Mix (Promega), 5 µL of cDNA, 50 µM of each S1OLIGO5’ and S1OLIGO3’ oligonucleotide [27] and nuclease-free water (Promega) in a final volume of 50 µL. The amplification conditions consisted of one cycle at 94 °C for one minute, followed by 35 cycles (94 °C for 30 s, 50 °C for 45 s, and 72 °C for 2 min) and a final extension at 72 °C for 10 min. The second PCR consisted of 25 µL of Go Taq® Green Master Mix (Promega), 2 µL of amplified DNA, 50 µM of each CK2 and CK4 oligonucleotide [24], and nuclease-free water (Promega) in a final volume of 50 µL. The amplification was carried out for one cycle at 94 °C for one minute, followed by 40 cycles (94 °C for 30 s, 50 °C for 45 s and 72 °C for 1 min) and a final step of 72 °C for 5 min. The PCR products separated by electrophoresis in a 1.5 % agarose gel together with a 100-bp DNA Ladder (Invitrogen™), stained with GelRed™ (Biotium) and viewed with the aid of a UV transilluminator. The amplification of the genomic target segment of IBV was performed in triplicate for each sample in the same run, and the products were sequenced separately.
Sequencing and analysis of the S1 gene sequence
The PCR products were sequenced by Macrogen Inc. (Seoul, Korea) and the contigs of the nucleotide sequences of the S1 gene were assembled using CLC Genomics Workbench version 7.5 (CLCbio). The partial sequences of the S1 gene were submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank), as shown in Table 1.
Table 1.
Information on Brazilian samples of infectious bronchitis virus (IBV) identified in two life periods of naturally infected chickens
| Strain | Isolation source | Isolation year | Accession number |
|---|---|---|---|
| Ma5 commercial vaccine | - | 2013 | KU736747 |
| UFV_bursa_fabricius/BR/13 | Chicks at 18 days | 2013 | KU736752 |
| UFV_intestine/BR/13 | Chicks at 18 days | 2013 | KU736753 |
| UFV_intestine_chicken/BR/14 | Hens at 364 days | 2014 | KU736754 |
| UFV_kidney/BR/13 | Chicks at 18 days | 2013 | KU736755 |
| UFV_kidney_chicken/BR/14 | Hens at 364 days | 2014 | KU736756 |
| UFV_embryonated_eggs/BR/14 | Chorioallantoic fluid | 2014 | KU736758 |
| UFV_lung/BR/13 | Chicks at 18 days | 2013 | KU736759 |
| UFV_oviduct_chicken/BR/14 | Hens at 364 days | 2014 | KU736760 |
| UFV_trachea/BR/13 | Chicks at 18 days | 2013 | KU736761 |
| UFV_cecal_tonsils_chicken/BR/14 | Hens at 364 days | 2014 | KU736762 |
BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to verify the identity of the sequences obtained in this study, using homologous sequences of IBV. After alignment of the sequences, an analysis was carried out of the polymorphisms found in the sequences using the MEGA application, version 6.0 [42].
A recent article has proposed a systematic IBV genotyping method in which a standard database of S1 nucleotide sequences is used to distinguish IBV strains [46]. Following this method, a database of the S1 gene was created that included the six partial sequences obtained in this study together with 14 sequences of previously described Brazilian isolates (BR-I and BR-II) and the 199 sequences described by Valastro et al. [46], as shown in a table in Online Resource 1. The 219 nucleotide sequences were aligned using the CLUSTALW algorithm. Phylogenetic analysis was performed using MEGA version 6 [42]. The sequences were grouped by the neighbor-joining distance method and Kimura’s two-parameter substitution model. The statistical support for the phylogenetic tree was calculated using 10,000 bootstrap replicates.
Results
Analysis of partial S1 sequences of IBV isolates obtained from chick samples
Samples that were positive for the S1 gene of IBV yielded a fragment of approximately 600 bp in nested RT-PCR. The nucleotide sequences of the S1 gene obtained in this study were identified directly from samples of 18-day-old chicks that showed clinical signs of IB and were compared with the sequence of the Ma5 vaccine (Table 2).
Table 2.
Polymorphism analysis of the partial S1 sequences from samples of chicks affected by the disease in relation to the sequence of the Ma5 vaccine. The polymorphisms are highlighted in bold and underlined
| Nucleotide position sample | 147 | 162 | 206 | 218 | 301 | 353 | 389 | 536 | 537 | 614 | 669 | 696 | 699 | 702 | 705 | 708 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ma5a | T | T | T | G | T | G | C | C | G | G | G | G | T | T | T | C |
| KU736761b | T | T | T | G | T | G | C | C | G | G | G | G | T | T | T | C |
| KU736753c | T | T | T | G | T | G | C | C | G | G | G | G | T | T | T | C |
| KU736759d | T | T | T | G | T | G | C | C | G | G | G | G | T | T | T | C |
| KU736752e | T | T | T | G | T | A | C | A | C | G | G | A | C | C | C | A |
| KU736755f | A | C | C | A | C | G | T | A | G | A | A | A | C | C | C | A |
The IBV samples identified in the trachea (KU736761), lung (KU736759), and small intestine (KU736753) showed 100 % identity to the Ma5 vaccine. However, in the bursa of Fabricius samples (KU736752), changes were observed in eight positions: G353A, C536A, G537C, G696A, T699C, T702C, T705C, and C708A. In the kidney samples (KU736755), the greatest variation was observed, totaling 14 changes: T147A, T162C, T206C, G218A, T301C, C389T, C536A, G614A, G669A, G696A, T699C, T702C, T705C, and C708A.
The similarity between the amino acids of the S1 protein of the chick samples was assessed between amino acids 29 and 218 of the pathogenic strain Mass 41 and the vaccine strain Ma5 (Table 3). Amino acid substitutions were observed in the bursa of Fabricius (KU736752) and kidney (KU736755) samples. In the sequence of the bursa of Fabricius sample, amino acid substitutions G100D and A161D were observed in relation to Ma5, and the changed amino acid at position 100 (D) was equivalent to the amino acid found in the pathogenic M41 strain. In the sequence of the kidney sample, amino acid substitutions I51T, G55D, Y83H, S112F, A161E, and R187K were observed in relation to MA5, and the changed amino acids at positions 51(T), 83 (H), 112 (F), 161 (E), and 187 (K) were identical to the amino acids of the pathogenic M41 strain. A substitution in the kidney sample was observed in hypervariable region 1 of the S1 gene (G55D).
Table 3.
Analysis of partial sequences of amino acids of the S1 gene from samples of chicks in relation to the Ma5 vaccine and the pathogenic strain Mass 41 (M21883). Amino acids that differ from those of the Ma5 vaccine are highlighted in bold. Amino acids that differ from those of Ma5 but are identical to those of the M41 strain are highlighted in bold and underlined. The locations of the two hypervariable regions follow the criteria defined by Kusters et al. [26]
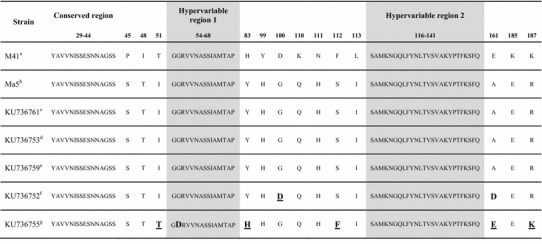
Analysis of a partial S1 sequence of IBV obtained from hen and embryo tissue samples
An S1 segment of IBV was amplified directly from the small intestine (KU736754), kidneys (KU736756), cecal tonsils (KU736762), and oviduct (KU736760) of hens and also from the chorioallantoic fluid of viable embryos after 18 days of incubation. The partial S1 sequences of IBV obtained from the intestine, kidneys, and oviduct of hens and from the chorioallantoic fluid of embryos were aligned with those of the Ma5 vaccine, and 100 % nucleotide sequence identity was observed.
In the cecal tonsils (KU736762), however, an IBV variant was detected that displayed significant genetic variation in relation to Ma5, as revealed by a total of 26.9 % of sites with polymorphisms along the entire sequence. In addition, amino acid insertions were observed in two distinct positions of the S1 gene, between codons 120 and 121 (KV) and between codons 143 and 147 (TGPSG), based on the amino acid sequence of the Ma5 strain (Table 4).
Table 4.
Analysis of partial sequences of amino acids of the S1 gene from hen and embryo samples. The amino acid sequence is based on the reference Ma5 strain (KU736747). The amino acid inserts in the S1 sequence of the IBV variants in the cecal tonsils (KU736762) are highlighted in bold
| Strain | Codons 120-121 | Codons 140-150 |
|---|---|---|
| Ma5a | --- | NGQ-----LFY |
| KU736754b | --- | NGQ-----LFY |
| KU736756c | --- | NGQ-----LFY |
| KU736760d | --- | NGQ-----LFY |
| KU736758e | --- | NGQ-----LFY |
| KU736762f | KV | QGNTGPSGLFY |
aMA5 vaccine (KU736747)
bIntestine
cKidney
dOviduct
eChorioallantoic fluid of embryonated eggs after 18 days of incubation
fCecal tonsils
Phylogenetic analysis
In order to construct a phylogenetic tree, the S1 IBV sequences were obtained directly from the clinical samples of the chicks (lung, KU736759; kidneys, KU736755; small intestine, KU736753; bursa of Fabricius, KU736752) and from the cecal tonsils of the hens at the end of the production cycle (KU736762), as well as from the Ma5 vaccine (KU736747), some previously described Brazilian isolates, and 199 reference strains (Fig. 1).
Fig. 1.
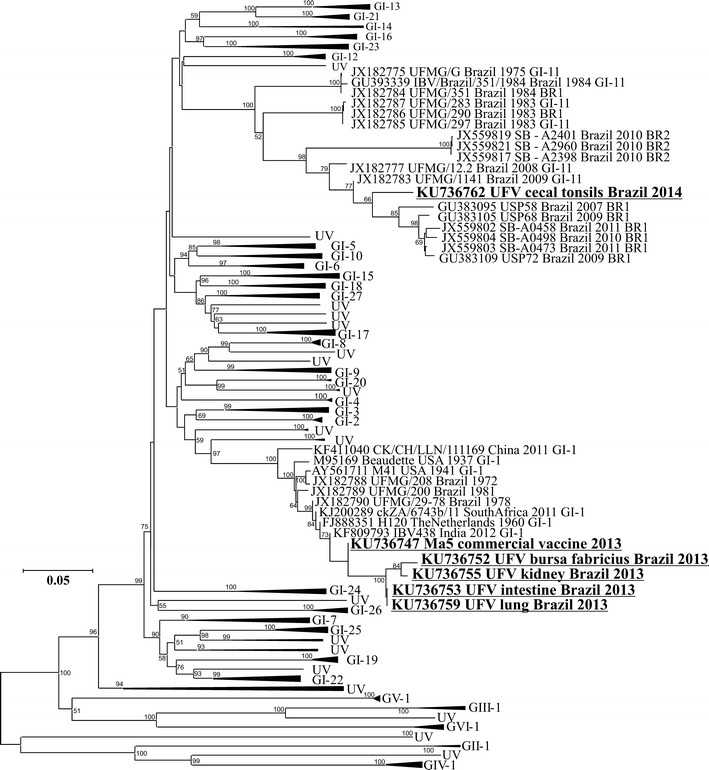
Phylogenetic analysis of 219 S1 sequences of IBV. The neighbor-joining phylogenetic tree shows the genetic relationship between the IBV strains identified directly from the clinical samples of the chicks (lung, KU736759; kidneys, KU736755; small intestine, KU736753; bursa of Fabricius, KU736752), from the cecal tonsils of the hens at the end of the production cycle (KU736762), and from the Ma5 vaccine (KU736747), some previously described Brazilian strains, and 199 reference strains. The numbers beside the nodes correspond to bootstrap values
As can be seen in the phylogenetic tree (Fig. 1), the viruses identified directly from the clinical samples of the chicks showed high similarity in the partial S1 sequence to the Ma5 vaccine and were therefore grouped with the GI-1 genotype (Massachusetts genotype) based on the method suggested by Valastro et al. [46]. The virus isolated from the cecal tonsils, on the other hand, showed a high degree of variation in the partial S1 sequence in comparison with the Ma5 vaccine, diverging from the Mass genotype, and it was therefore grouped with other strains that were previously characterized as belonging to the Brazilian variant genotype (GI-11).
Discussion
In this study, we analyzed the genetic diversity of IBV at the field level in a batch of breeding hens that had been vaccinated with the Ma5 vaccine and naturally affected by IB in early life. Furthermore, the molecular dynamics and the vertical transmission of IBV in the same batch of hens at the end of the production cycle were investigated. IBV was identified in different tissues, including the bursa of Fabricius, kidney, trachea, lung, and small intestine from the chick samples. IBV variants identified in the kidney and bursa of Fabricius in the chick samples were not detected in hens, but another IBV variant was identified in the cecal tonsils. In addition, vertical transmission could be observed through the identification of IBV in the chorioallantoic fluid of live embryonated eggs.
It was demonstrated that chicks vaccinated at one day of age did not have enough protection compared with chicks vaccinated at 14 days of age [44]. Another study has pointed out that an efficient immune response with the use of specific IBV vaccines in the field limits viral replication and restricts the generation of genetically distinct strains in the same host [35]. Although the chicks examined in this study were vaccinated, early immunization seems to be influenced by maternal antibodies, resulting in a failure of the Ma5 vaccine to provide protection, and consequently, additional opportunities for the generation of genetic viral diversity in the infected chickens.
In this study, we observed that the partial S1 sequences of IBV obtained from the bursa of Fabricius and kidneys of clinical samples exhibited genetic differences in relation to the Ma5 vaccine (Tables 2 and 3). Additionally, the S1 sequence associated with the kidney sample had a higher degree of variation than the virus identified in the bursa of Fabricius. In agreement with the literature [3, 4, 36], we suggest that the disease affecting the chicks at the 18th day of life was caused primarily by a nephropathogenic strain (KU736755).
Differences in tissue tropism and thus in the pathogenicity of IBV strains have been hypothesized to be associated with differences in their spike (S) proteins [5, 48]. We suggest that the amino acid sites 51, 83, 112, 161, and 187 of the S1 protein are involved in the pathogenicity of IBV strains, because changes in these amino acids detected in the nephropathogenic strain (KU736755) are identical to the amino acids found in the pathogenic strain M41.
It has been shown that the strains M41 and Beaudette-US use sialic acid for adsorption [49], and infection of the tracheal epithelium is dependent on sialic acid [50]. Recently, the receptor-binding domain of the IBV M41 strain was mapped to residues 16–69 in the S1 protein [40]. Our analysis of partial sequences of amino acids of the S1 gene showed that changes in the amino acids of the strains obtained from the kidneys were located at residues 51, 55, 83, 113, 161, and 187. The receptor-binding domain of the IBV (aa. 16-69) includes the 51-55 region identified in this study. These results suggest that the changes found in this region can be correlated with the pathogenicity of the nephropathogenic strain (KU736755).
While the ability to bind to susceptible host cells is the first step in viral infection, the host innate immune response is also a major contributing factor to the pathological outcome of IBV infection [9]. The relationship between mutations, tropism and pathogenicity are complex and involve both genetic characteristics of IBV and the innate immune response mounted by the host.
In the samples of small intestine, kidneys and oviduct from the hens of this batch that reached the end of production (52 weeks of age), segments of S1 with 100 % identity to those of the Ma5 vaccine were identified. However, IBV field strains identified in the kidney and bursa of Fabricius in the chick samples were not detected in hens at the end of the production cycle. This fact suggests that the elimination of these strains is associated with multiple factors inherent in the host that are responsible for the natural selection process, such as the adaptive immune response acquired after vaccination of the batch [43].
Studies have shown the persistence of IBV strains at the experimental level [1, 10, 11, 13, 38]. We detected S1 segments with 100 % identity to the Ma5 vaccine in the small intestine, kidneys, and oviduct of hens at the end of the production cycle. This result suggests that the S1 fragments detected in the hens are evidence of persistence of the Ma5 vaccine.
The phenomenon of persistence of IBV strains, including vaccine strains, may have an impact on the generation of genetic diversity due to the high frequency of RNA recombination in coronaviruses [23]. Sequencing of many field strains has provided convincing evidence that many, possibly all, IBV strains are recombinants between different field strains [7, 18, 25, 30] as well as between field strains and strains used in live vaccines [28, 29].
An IBV strain was also identified in the cecal tonsils. The virus identified in the cecal tonsils exhibited considerable variation in the partial S1 sequence in relation to the Ma5 vaccine and other strains detected in the chicks. Due to the genetic variation displayed by this virus, it was grouped in the phylogenetic tree with the previously described genotype BR-I [8, 15, 47] or GI-11 [46]. Brazilian IBV variants are usually identified in the cecal tonsil and have already been grouped into the two genotypes BR-I and BR-II [8, 15, 47].
Consistent with the findings of this work, the vertical transmission of IBV in hens has been reported in an outbreak of IB. On the experimental level, IBV has been identified in the eggs of challenged hens, as well as in one-day-old chicks [12, 33]. In this work, the detection of the S1 segment in the oviduct and viable embryos with 100 % identity to that of the Ma5 vaccine possibly reflects the vertical transmission and the nonpathogenic nature of the vaccinal virus. On the other hand, we cannot exclude the possibility of vertical transmission of the virus variant identified in the cecal tonsils to the embryonated egg.
The distribution of genetically distinct strains in different tissues demonstrates the potential for genetic variation that an IBV infection may cause in naturally infected chickens. In addition, vertical transmission is an important factor in the control of IB, but this phenomenon seems to have been neglected. These findings prove to be relevant because they provide information about the genetic diversity of IBV in a natural infection that can be applied to immunoprophylaxis programs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was supported by the Brazilian Government Agencies CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais).
Compliance with ethical standards
The authors declare no conflicts of interest. This project complied with the principles of the Commission for Ethics in Animal Experimentation of the Universidade Federal de Viçosa (UFV) under protocol no. 74/2013. All authors contributed to this work and agreed to its publication.
References
- 1.Alexander DJ, Gough RE. A long-term study of the pathogenesis of infection of fowls with three strains of avian infectious bronchitis virus. Res Vet Sci. 1978;24:228–233. [PubMed] [Google Scholar]
- 2.Almeida DO, Tortelly R, Nascimento ER, Chagas MA, Khan MI, Pereira VL. Avian infectious bronchitis and deep pectoral myopathy—a case control study. Poultry Sci. 2012;91:3052–3056. doi: 10.3382/ps.2012-02476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awad F, Baylis M, Ganapathy K (2014) Detection of variant infectious bronchitis viruses in broiler flocks in Libya. IJVSM 2(2):78–82. doi:10.1016/j.ijvsm.2014.01.001
- 4.Benyeda Z, Mató T, Süveges T, Szabó E, Kardi V, Abonyi-Tóth Z, Rusvai M, Palya V. Comparison of the pathogenicity of QX-like, M41 and 793/B infectious bronchitis strains from different pathological conditions. Avian Pathol. 2009;38:449–456. doi: 10.1080/03079450903349196. [DOI] [PubMed] [Google Scholar]
- 5.Casais R, Dove B, Cavanagh D, Britton P. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J Virol. 2003;77:9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh D (2005) Coronaviruses in poultry and other birds. Avian Pathol 34:439–448. doi:10.1080/03079450500367682 [DOI] [PubMed]
- 7.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 8.Chacón JL, Rodrigues JN, Assayag Junior MS, Peloso C, Pedroso AC, Ferreira AJ. Epidemiological survey and molecular characterization of avian infectious bronchitis virus in Brazil between 2003 and 2009. Avian Pathol. 2011;40:153–162. doi: 10.1080/03079457.2010.544641. [DOI] [PubMed] [Google Scholar]
- 9.Chhabra R, Kuchipudi SV, Chantrey J, Ganapathy K. Pathogenicity and tissue tropism of infectious bronchitis virus is associated with elevated apoptosis and innate immune responses. Virology. 2016;488:232–241. doi: 10.1016/j.virol.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong KT, Apostolov K. The pathogenesis of nephritis in chickens induced by infectious bronchitis virus. J Comp Pathol. 1982;92:199–211. doi: 10.1016/0021-9975(82)90078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook JK. Duration of experimental infectious bronchitis in chickens. Res Vet Sci. 1968;9:506–514. [PubMed] [Google Scholar]
- 12.Cook JKA. Recovery of infectious bronchitis virus from eggs and chicks produced by experimentally inoculated hens. J Comp Pathol. 1971;81:203–211. doi: 10.1016/0021-9975(71)90093-4. [DOI] [PubMed] [Google Scholar]
- 13.El-Houadfi M, Jones RC, Cook JKA, Ambali AG. The isolation and characterisation of six avian infectious bronchitis viruses isolated in Morocco. Avian Pathol. 1986;15:93–105. doi: 10.1080/03079458608436269. [DOI] [PubMed] [Google Scholar]
- 14.Feng J, Hu Y, Ma Z, Yu Q, Zhao J, Liu X, Zhang G. Virulent avian infectious bronchitis virus, People’s Republic of China. Emerg Infect Dis. 2012;18:1994–2001. doi: 10.3201/eid1812.120552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraga AP, Balestrin E, Ikuta N, Fonseca AS, Spilki FR, Canal CW, Lunge VR. Emergence of a new genotype of avian infectious bronchitis virus in Brazil. Avian Dis. 2013;57:225–232. doi: 10.1637/10346-090412-Reg.1. [DOI] [PubMed] [Google Scholar]
- 16.Gallardo RA, van Santen VL, Toro H. Host intraspatial selection of infectious bronchitis virus populations. Avian Dis. 2010;54:807–813. doi: 10.1637/9054-090809-Reg.1. [DOI] [PubMed] [Google Scholar]
- 17.Gallardo RA, Hoerr FJ, Berry WD, van Santen VL, Toro H. Infectious bronchitis virus in testicles and venereal transmission. Avian Dis. 2011;55:255–258. doi: 10.1637/9592-102910-Reg.1. [DOI] [PubMed] [Google Scholar]
- 18.Hewson KA, Ignjatovic J, Browning GF, Devlin JM, Noormohammadi AH. Infectious bronchitis viruses with naturally occurring genomic rearrangement and gene deletion. Arch Virol. 2011;156:245–252. doi: 10.1007/s00705-010-0850-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong SM, Kwon HJ, Kim IH, Mo ML, Kim JH. Comparative genomics of Korean infectious bronchitis viruses (IBVs) and an animal model to evaluate pathogenicity of IBVs to the reproductive organs. Viruses. 2012;4:2670–2683. doi: 10.3390/v4112670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackwood MW, Hilt DA, Lee CW, Kwon HM, Callison SA, Moore KM, Moscoso H, Sellers H, Thayer S. Data from 11 years of molecular typing infectious bronchitis virus field isolates. Avian Dis. 2005;49:614–618. doi: 10.1637/7389-052905R.1. [DOI] [PubMed] [Google Scholar]
- 21.Jackwood MW, Hilt DA, McCall AW, Polizzi CN, McKinley ET, Williams SM. Infectious bronchitis virus field vaccination coverage and persistence of Arkansas-type viruses in commercial broilers. Avian Dis. 2009;53:175–183. doi: 10.1637/8465-090308-Reg.1. [DOI] [PubMed] [Google Scholar]
- 22.Jackwood MW, Hall D, Handel A. Molecular evolution and emergence of avian gammacoronaviruses. Infect Genet Evol. 2012;12:1305–1311. doi: 10.1016/j.meegid.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong YS, Repass JF, Kim YN, Hwang SM, Makino S. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology. 1996;217:311–322. doi: 10.1006/viro.1996.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keeler CL, Jr, Reed KL, Nix WA, Gelb J., Jr Serotype identification of avian infectious bronchitis virus by RT-PCR of the peplomer (S-1) gene. Avian Dis. 1998;42:275–284. doi: 10.2307/1592477. [DOI] [PubMed] [Google Scholar]
- 25.Kuo SM, Kao HW, Hou MH, Wang CH, Lin SH, Su HL. Evolution of infectious bronchitis virus in Taiwan: positively selected sites in the nucleocapsid protein and their effects on RNA-binding activity. Vet Microbiol. 2013;162:408–418. doi: 10.1016/j.vetmic.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusters JG, Niesters HG, Lenstra JA, Horzinek MC, van der Zeijst BA. Phylogeny of antigenic variants of avian coronavirus IBV. Virology. 1989;169:217–221. doi: 10.1016/0042-6822(89)90058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon HM, Jackwood MW, Gelb J., Jr Differentiation of infectious bronchitis virus serotypes using polymerase chain reaction and restriction fragment length polymorphism analysis. Avian Dis. 1993;37:194–202. doi: 10.2307/1591474. [DOI] [PubMed] [Google Scholar]
- 28.Lim T, Lee HJ, Lee DH, Lee YN, Park JK, Youn HN, Kim MS, Lee JB, Park SY, Choi IS, Song CS. An emerging recombinant cluster of nephropathogenic strains of avian infectious bronchitis virus in Korea. Infect Genet Evol. 2011;11:678–685. doi: 10.1016/j.meegid.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Ma H, Xu Q, Sun N, Han Z, Sun C, Guo H, Shao Y, Kong X, Liu S. Characterization of a recombinant coronavirus infectious bronchitis virus with distinct S1 subunits of spike and nucleocapsid genes and 3’ untranslated region. Vet Microbiol. 2013;162:429–436. doi: 10.1016/j.vetmic.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Xu Q, Han Z, Liu X, Li H, Guo H, Sun N, Shao Y, Kong X. Origin and characteristics of the recombinant novel avian infectious bronchitis coronavirus isolate ck/CH/LJL/111054. Infect Genet Evol. 2014;23:189–195. doi: 10.1016/j.meegid.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucio B, Fabricant J. Tissue tropism of three cloacal isolates and Massachusetts strain of infectious bronchitis virus. Avian Dis. 1990;34:865–870. doi: 10.2307/1591375. [DOI] [PubMed] [Google Scholar]
- 32.Matthijs MG, van Eck JH, Landman WJ, Stegeman JA. Ability of Massachusetts-type infectious bronchitis virus to increase colibacillosis susceptibility in commercial broilers: a comparison between vaccine and virulent field virus. Avian Pathol. 2003;32:473–481. doi: 10.1080/0307945031000154062. [DOI] [PubMed] [Google Scholar]
- 33.McFerran JB, Cahill HT, Young JA, Wright CL. Isolation of infectious bronchitis virus from newborn chicks and dead-in-shell embryos. Vet Rec. 1971;89:560–561. doi: 10.1136/vr.89.21.560. [DOI] [PubMed] [Google Scholar]
- 34.McKinley ET, Hilt DA, Jackwood MW. Avian coronavirus infectious bronchitis attenuated live vaccine undergo selection of subpopulations and mutations following vaccination. Vaccine. 2008;26:1274–1284. doi: 10.1016/j.vaccine.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKinley ET, Jackwood MW, Hilt DA, Kissinger JC, Robertson JS, Lemke C, Paterson AH. Attenuated live vaccine usage affects accurate measures of virus diversity and mutation rates in avian coronavirus infectious bronchitis virus. Virus Res. 2011;158:225–234. doi: 10.1016/j.virusres.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meir R, Rosenblut E, Perl S, Kass N, Ayali G, Perk S, Hemsani E. Identification of a novel nephropathogenic infectious bronchitis virus in Israel. Avian Dis. 2004;48:635–641. doi: 10.1637/7107. [DOI] [PubMed] [Google Scholar]
- 37.Mork AK, Hesse M, Abd El Rahman S, Rautenschlein S, Herrler G, Winter C. Differences in the tissue tropism to chicken oviduct epithelial cells between avian coronavirus IBV strains QX and B1648 are not related to the sialic acid binding properties of their spike proteins. Vet Res. 2014;14:45–67. doi: 10.1186/1297-9716-45-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naqi S, Gay K, Patalla P, Mondal S, Liu R. Establishment of persistent avian infectious bronchitis virus infection in antibody-free and antibody-positive chickens. Avian Dis. 2003;47:594–601. doi: 10.1637/6087. [DOI] [PubMed] [Google Scholar]
- 39.Ndegwa EN, Toro H, van Santen VL. Comparison of vaccine subpopulation selection, viral loads, vaccine virus persistence in trachea and cloaca, and mucosal antibody responses after vaccination with two different Arkansas Delmarva poultry industry-derived infectious bronchitis virus vaccines. Avian Dis. 2014;58:102–110. doi: 10.1637/10609-070613-Reg.1. [DOI] [PubMed] [Google Scholar]
- 40.Promkuntod N, van Eijndhoven RE, de Vrieze G, Gröne A, Verheije MH. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. 2014;448:26–32. doi: 10.1016/j.virol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raj GD, Jones RC. Infectious bronchitis virus: immunopathogenesis of infection in the chicken. Avian Pathol. 1997;26:677–706. doi: 10.1080/03079459708419246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toro H, van Santen VL, Jackwood MW. Genetic diversity and selection regulates evolution of infectious bronchitis virus. Avian Dis. 2012;56:449–455. doi: 10.1637/10072-020212-Review.1. [DOI] [PubMed] [Google Scholar]
- 44.van Ginkel FW, Padgett J, Martinez-Romero G, Miller MS, Joiner KS, Gulley SL. Age-dependent immune responses and immune protection after avian coronavirus vaccination. Vaccine. 2015;33:2655–2661. doi: 10.1016/j.vaccine.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Santen VL, Toro H. Rapid selection in chickens of subpopulations within ArkDPI-derived infectious bronchitis virus vaccines. Avian Pathol. 2008;37:293–306. doi: 10.1080/03079450802043783. [DOI] [PubMed] [Google Scholar]
- 46.Valastro V, Holmes EC, Britton P, Fusaro A, Jackwood MW, Cattoli G, Monne I. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect Genet Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villarreal LY, Sandri TL, Souza SP, Richtzenhain LJ, de Wit JJ, Brandao PE. Molecular epidemiology of avian infectious bronchitis in Brazil from 2007 to 2008 in breeders, broilers, and layers. Avian Dis. 2010;54:894–898. doi: 10.1637/9218-121709-Reg.1. [DOI] [PubMed] [Google Scholar]
- 48.Wickramasinghe IN, deVries RP, Grone A, de Haan CA, Verheije MH. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J Virol. 2011;85:8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winter C, Schwegmann-Wessels C, Cavanagh D, Neumann Herrler G. Sialic acid is a receptor determinant for infection of cells by avian infectious bronchitis virus. J Gen Virol. 2006;87:1209–1216. doi: 10.1099/vir.0.81651-0. [DOI] [PubMed] [Google Scholar]
- 50.Winter C, Herrler G, Neumann U. Infection of the tracheal epithelium by infectious bronchitis virus is sialic acid dependent. Microbes Infect. 2008;10:367–373. doi: 10.1016/j.micinf.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


