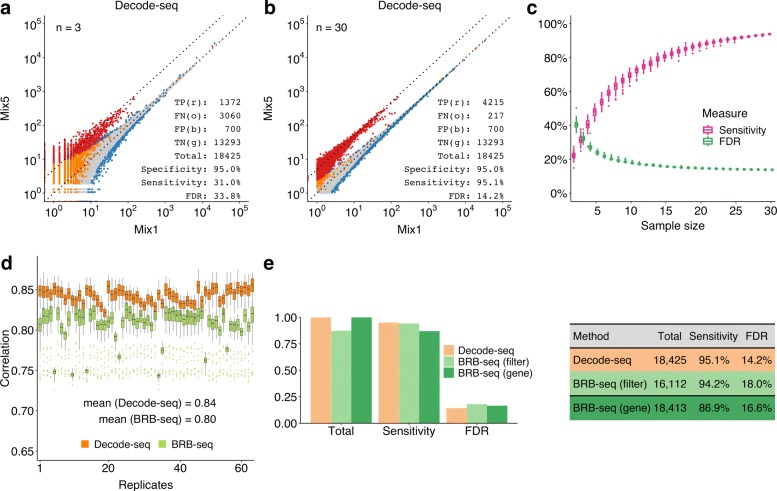
Fig. 1.
Performance evaluation of Decode-seq. a Differential expression analysis with 3 pairs of replicates. True positive (TP, red dots), mouse genes which were called DE; false negative (FN, orange dots), mouse genes which were called non-DE; true negative (TN, gray dots), human genes which were called non-DE; false positive (FP, blue dots), human genes which were called DE. Specificity = TN/(TN + FP), and it was fixed to 95% in all calculation. Sensitivity = TP/(TP + FN). False discovery rate = FP/(TP + FP). b DE analysis with 30 pairs of replicates. The sensitivity increased to 95.1%, and the false discovery rate dropped to 14.2%. c DE performance related to replicate number calculated by random downsampling of 30-pair data. Each replicate number was calculated 100 times. Sensitivity and false discovery rate were improved dramatically when the number of replicates increased. d Spearman’s correlations among replicates of Decode-seq and BRB-seq. Each replicate was compared with all other replicates of Decode-seq and BRB-seq, respectively. The distribution of these correlations was shown as the box for each replicate. In the Decode-seq group, replicate has higher correlations with each other, indicating the higher reproducibility. e DE performance of Decode-seq and BRB-seq. Bars in three colors represent DE performance of three sets: Decode-seq, BRB-seq with the same filter parameters as Decode-seq, and BRB-seq with the same total gene as Decode-seq. When using the same filter parameters, BRB-seq detected fewer genes. When using loose filter parameters to ensure the same total gene, BRB-seq gave a lower sensitivity