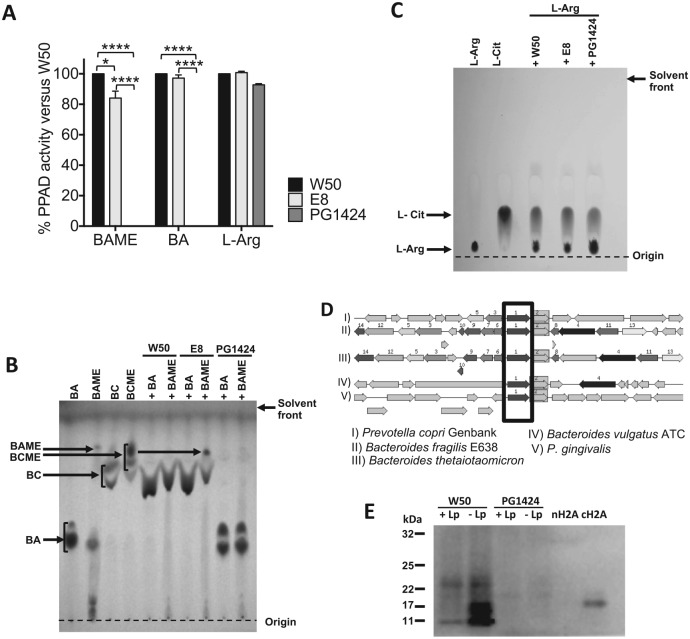
Figure 1.
Porphyromonas gingivalis peptidylarginine deiminase (PPAD) activity can be detected in the wild-type but not the PPAD-mutant P. gingivalis strain. (A) Percentage of PPAD activity in P. gingivalis E8 (rgpA-rgpB mutant) and PG1424 (ppad mutant) in comparison with P. gingivalis W50 (wild-type strain) measured by colorimetric assay. Results are expressed as mean ± SEM. *P < 0.05. ****P < 0.0001. One-way analysis of variance, followed by Tukey’s multiple- comparisons test on the raw data. BA, N-α-benzoyl-L-arginine; BAME, N-α-benzoyl-L-arginine methyl ester; BC, N-α-benzoyl L-citrulline; BCME, N-α-benzoyl L-citrulline methyl ester. (B) BC and BCME produced by P. gingivalis strains W50, E8, and PG1424 after incubation with BA or BAME were measured by thin layer chromatography. (C) L-citrulline production from L-arginine by P. gingivalis strains W50, E8, and PG1424 was assessed by thin layer chromatography. (D) Sequence comparisons of the P. gingivalis genome with arginine deiminases (in box) of other bacterial species to identify orthologous protein sequences. (E) Comparison of autocitrullination in P. gingivalis W50 and PG1424. Immunoblotting of whole cell lysates from P. gingivalis W50 and PG1424 prepared with immediate addition of leupeptin (+Lp) or delayed addition (–Lp; after 10 min of lysis) was carried out via the antimodified citrulline method, including native histone H2A (nH2A; negative control) and citrullinated histone H2A (cH2A; positive control).