Fig. 4.
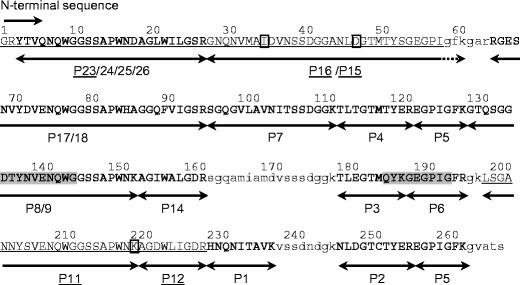
The predicted amino acid sequence of native KAA-2. The results of sequence prediction by peptide mapping in KAA-2 are shown using ESA-2 sequence as a template. The peptide fragments produced by trypsin digestion of PE-KAA-2 are represented with arrows. The peak numbers examined by Edman degradation and their determined sequences are underlined. The sequences whose peptide masses were consistent with those of ESA-2 are represented in bold. Unidentified amino acids in KAA-2 are represented as lower case letter of corresponding sequence in ESA-2. Shaded regions in the sequence were used to design the degenerated primers for cDNA cloning
