Abstract
Both, the influenza C (INF-C) virus haemagglutinin esterase fusion and bovine coronavirus (BCoV) haemagglutinin esterase surface glycoproteins exhibit a lectin binding capability and a receptor-destroying 9-O-acetyl esterase activity that recognise 9-O-acetyl-N-acetylneuraminic acid (Neu5,9Ac2)-containing glycans. Here we report nuclear magnetic resonance and molecular modelling studies on the 9-O-acetyl esterase showing that the α-configured Neu5,9Ac2 is strictly preferred by the INF-C and BCoV esterases. Interestingly, we have discovered that the INF-C esterase function releases acetate independently of the chemical nature of the aglycon moiety, whereas subtle differences in substrate recognition were found for BCoV esterase. Analysis of the apo and complexed X-ray crystal structure of INF-C esterase revealed that binding of 9-O-acetylated N-acetylneuraminic acids is a dynamic process that involves conformational rearrangement of serine-57 in the esterase active site. This study provides valuable insights towards the design of drugs to combat INF-C virus and coronavirus infections causing outbreaks of upper respiratory infections and severe diarrhea in calves, respectively.
Keywords: Influenza C virus esterase, Bovine coronavirus esterase, NMR spectroscopy, Molecular modeling
Introduction
Influenza C (INF-C) virus causes outbreaks of upper respiratory infections worldwide and the risk of complications with lower-respiratory-tract illness is high in children younger than 2 years of age [1]. In contrast to influenza A and B viruses, which bind to N-acetylneuraminic acid (Neu5Ac, 1) [2, 3], INF-C viruses strictly employ 9-O-acetylated N-acetylneuraminic acid (9-O-acetyl-N-acetylneuraminic acid, Neu5,9Ac2, 2) as a receptor [4, 5] (Fig. 1). The INF-C haemagglutinin esterase fusion (HEF) protein serves as a receptor-binding protein, displays receptor-destroying 9-O-acetyl esterase activity [6, 7] and promotes fusion of the viral and endosomal membranes [5]. Neu5,9Ac2 (2) occurs ubiquitously also on non-suitable host cells, on virions and on soluble and insoluble molecules in the extracellular environment. The esterase activity provides an escape ticket for the virion progeny preventing irreversible non-productive binding to such receptors. Besides INF-C viruses, infectious salmon anaemia virus [8, 9], toroviruses [10] and group 2 coronaviruses [11–17] express evolutionarily related O-acetylesterases. Bovine coronavirus (BCoV) is closely related to the human respiratory coronaviruses HCoV-OC43 and HKU [18, 19] and causes severe diarrhoea in calves and winter dysentery in adult cattle resulting in loss of productivity in the livestock industry. Viral sialate-O-acetylesterases represent a family of serine (Ser) hydrolases [20]. They covalently bind the serine hydrolase inhibitor diisopropyl-fluorophosphate (DFP) [8, 12, 21]. By labeling with 3H-DFP, the active site serine residue of the INF-C virus esterase was identified as serine-71 of the precursor protein [20, 22], which after proteolytic removal of the signal peptide corresponds to serine-57 in the mature HEF protein. The active site of serine esterases is composed of a charge relay system [23], also termed catalytic triad. In the mature INF-C HEF protein this triad is formed by Ser-57, histidine (His)-355 and aspartate (Asp)-352 [24].
Fig. 1.
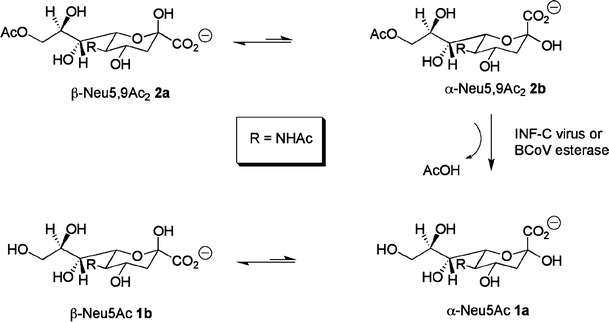
De-O-acetylation of Neu5,9Ac2 by INF-C or BCoV esterase
To gain a better understanding of the mechanism and mode of action of O-acetylesterases, nuclear magnetic resonance (NMR) studies in combination with molecular modelling studies of the receptor-destroying 9-O-acetyl esterase from INF-C virus and BCoV (Scheme 1) have been undertaken. Specifically, we have utilized one-dimensional 1H NMR spectroscopy to follow the enzyme-catalyzed reaction of the INF-C and BCoV esterases using a number of 9-O-acetyl N-acetylneuraminic acid derivatives. NMR-based enzyme assays allowed simultaneous monitoring of the consumption of substrates and subsequent build-up of products. Our NMR and molecular modelling experiments show that the Neu5,9Ac2 in its α-configuration is strictly preferred by both the INF-C and BCoV esterase. Interestingly, the INF-C esterase function releases acetate completely independently of the chemical nature of the aglycon moiety.
Materials and methods
Expression of influenza C virus HEF and bovine coronavirus HE
The INF-C virus HEF used in this study comprises of a chimeric recombinant soluble influenza C/Cal/78 virus haemagglutinin esterase protein fused in frame to enhanced green fluorescent protein (HE12-GFP). The HE12-GFP protein was expressed in insect Sf9 cells in serum-free media by recombinant baculoviruses [25]. The HE12-GFP was purified from cell culture supernatant by ultracentrifugation followed by an application to a Q-Sepharose FF column. The esterase was dialyzed against 25 mM Na-phosphate pH 7.0 and freeze-dried for transport. The lyophilized enzyme was resolved in deuterated phosphate buffer as used for NMR analysis. The esterase activity of INF-C virus HEF and coronavirus HE was determined with p-nitrophenyl acetate (pNPA) as described previously [7, 25]. One milliunit of viral esterase activity was defined as the amount of enzymatic activity resulting in the hydrolysis of 1 nmol of pNPA per minute. The ectodomain (residues 19–388) of BCoV strain Mebus was expressed as an Fc fusion protein in human embryonic kidney 293 cells as described previously [26] and purified from the tissue culture supernatant by protein A-affinity chromatography.
Preparation of 9-O-acetylated N-acetylneuraminides
Figure 2 shows the 9-O-acetylated N-acetylneuraminides used in this study. Neu5,9Ac2 (2) was isolated from bovine submaxillary mucin by acid hydrolysis [27] and purified as previously described [15] followed by a second purification with a Dowex 1 × 8 (mesh 200–400) anion exchange column. The known 9-O-acetylated methyl N-acetylneuraminides Neu5,9Ac2α2Me (3) [28], Neu5,9Ac2β2Me (5) [29], and thio-linked N-acetylneuraminosylglucoside, Neu5,9Ac2α(2-6)-S-Glcβ1Me (4) [30] were readily synthesized from the corresponding N-acetylneuraminide in 80%, 85% and 70% isolated yields respectively according to published protocols.
Fig. 2.
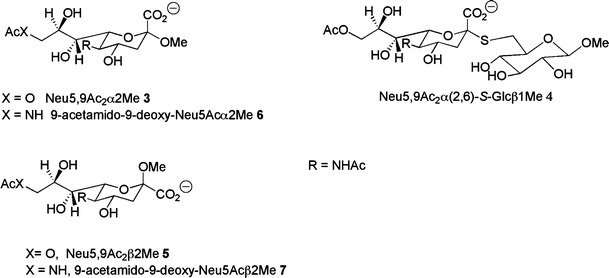
Structures of C9-functionalised N-acetylneuraminides
NMR experiments
NMR-based INF-C virus and BCoV esterase assays were analysed on a Bruker 600 UltraShield™ at 298 K in deuterated 100 mM phosphate buffer, 100 mM NaCl, pH 7.0. 1H NMR spectra were acquired with 32 scans, a 2 s relaxation delay over a spectral width of 6,000 Hz. Solvent suppression of the residual HDO peak was achieved by continuous low power presaturation pulse during the relaxation delay. Data acquisition and processing were performed with XWINNMR software. Substrates 2, 3, 4, and 5 were dissolved in NMR buffer and a 1 mM solution in a total volume of 600 μL in the absence of protein was prepared and a 1H NMR spectrum acquired. After addition of 20 mU HE12-GFP and 6.6 mU BCoV 1H NMR spectra were recorded in 5 min intervals over 100 min (Fig. 3). An identical protocol was used to follow the reaction of BCoV esterase (Fig. 3). Prior to NMR-based enzyme assay chemical shift assignment of all used substrates was achieved by correlation spectroscopy, total correlation spectroscopy, nuclear overhauser enhancement spectroscopy and heteronuclear single quantum correlation NMR experiments.
Fig. 3.
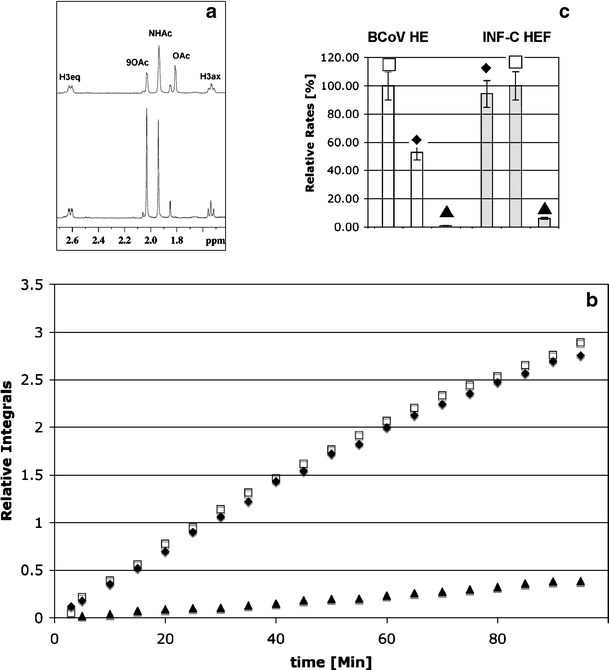
a 600 MHz 1H NMR spectrum of Neu5,9Ac2 α2Me (3) at 0 min (bottom panel) and after 60 min incubation with INF-C virus HEF (top panel); b relative integrals of the acetate 1H NMR signal (1.81 ppm) after INF-C virus HEF incubation; c comparison of the relative rates of the acetate release by INF-C virus HEF and BCoV HE. The following substrates were used: Neu5,9Ac2α2Me (3; square), Neu5,9Ac2α(2,6)-S-Glcβ1Me (4; filled diamond), and Neu5,9Ac2 (2; filled triangle)
Molecular modelling and forced torsion grid search
Molecular modelling investigation was performed with the crystal structure [31] (Protein Data Bank acc. no: 1FLC) and the crystal structure pro and visualized with Insight II (Accelrys. InsightII, Accelrys Inc., San Diego, CA, USA 2001) software. The forced torsion grid search used carried out by the extraction of the Neu5,9Ac2 structure from the crystal structure [31] and used as the starting point to build Neu5,9Ac2α2-6-S-Glcβ1Me using InsightII. The three torsion angles involved in the 2-6-glycosidic linkage (Φ,Ψ,Ω) were forcibly rotated using Discover (Discover, Accelrys Inc., San Diego, CA, USA 2001), in increments of 90° with the intramolecular energy being evaluated at each step using consistent valence force field [32]. Conformations that were within 25 kcal/mol of the lowest energy structure were extracted and viewed in InsightII using a biosym command language macro that superimposed the ring atoms of the sialic acid of each conformation onto the ring atoms of the sialic acid in the crystal structure. The results are viewed in Fig. 4 where conformational flexibility is displayed relative to the solvent accessible surface of the HEF active site.
Fig. 4.
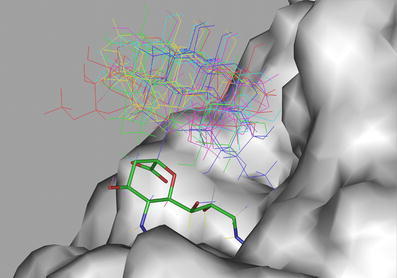
Superimposition of the sialic acid portion of a set of conformations of Neu5,9Ac2α(2–6)-S-Glcβ1Me (4) with the methyl 9-acetamido-9-deoxy-N-acetyl-α-D-neuraminide (9-acetamido-9-deoxy-Neu5Acα2Me, 6) within the active site of INF-C HEF. The glucose moiety of 6 does not interact with the protein surface and more likely has no effect on acetate cleavage and release
Results and discussion
Figure 3a shows the 1H NMR spectrum of Neu5,9Ac2α2Me (3) after incubation of with INF-C HEF for 1 h. A dramatic reduction of the 9-O-acetyl signal (9-O-Ac, Fig. 3a; intensity at 2.05 ppm) and simultaneously an increase of the intensity of the free acetate signal (Fig. 3a) at 1.81 ppm could be detected. No changes in intensity of the 1H NMR signal of the methyl protons of the N-acetamido moiety (1.92 ppm) could be observed, because de-O-acetylation at C9 has no effect on the chemical shift of the N-acetamido group at C5 (Fig. 3a). To study the activity of INF-C HEF towards different substrates and to determine the effect of aglycon groups, we compared the time-based acetate release for Neu5,9Ac2α2Me (3), Neu5,9Ac2α(2-6)-S-Glcβ1Me (4), and Neu5,9Ac2 (2; Fig. 3b). INF-C HEF de-O-acetylated all three substrates, but Neu5,9Ac2 (2) only inefficiently. Interestingly, Neu5,9Ac2α2Me (3) and Neu5,9Ac2α(2-6)-S-Glcβ1Me (4) were cleaved at similar rates. We conclude that for these substrates the rate of acetate release is independent of the chemical composition of the aglycon moiety (methyl and sulfur α(2-6)-linked glucose unit, respectively). To provide further evidence that the acetate release is independent of the chemical composition of the aglycon unit, we have re-examined the x-ray crystal structure [24, 30] of INF-C virus HEF in complex with the α-methyl glycoside of 9-acetamido-9-deoxy-N-acetylneuraminic acid (9-acetamido-9-deoxy-Neu5Acα2Me, 6). The 9-O-acetyl sialic acid portion of the disaccharide 4 was superimposed with the monosaccharide 6. Sixty-four different conformations of the thio-α(2-6)-linkage were generated to access an ensemble of conformations of four. Figure 4 illustrates low-energy conformations of 4 superimposed with 6 within the active site of INF-C HEF. It is immediately obvious that in these selected conformations, the glucose unit appears to have little, if any, interaction with the protein surface. This observation is evidence that the aglycon moiety does not interact with the protein and, more importantly, not with the acetate cleavage site, supporting our assumption that the rate of acetate release for INF-C HEF is independent of the chemical composition of the aglycon moiety.
We reasoned that the reduced enzymatic activity of HEF towards Neu5,9Ac2 (2) might be related to the solution anomeric ratio of this substrate. Both Neu5,9Ac2α2Me (3) and Neu5,9Ac2α(2-6)-S-Glcβ1Me (4) are locked in the α-configuration whereas Neu5,9Ac2 (2) has an α:β anomeric ratio of ~5:95, as determined by 1H NMR spectroscopy (data not shown). To investigate this directly, we measured acetate release from the strictly β-anomerically-configured substrate, Neu5,9Ac2β2Me (5). Our NMR-based enzyme assay revealed that after 1 h incubation with INF-C HEF, the 9-O-acetyl signal (2.05 ppm) had the same intensity compared to the 1H NMR spectrum in the absence of the enzyme. From this observation, we conclude that the β-anomer does not interact with the HEF esterase in an optimal manner. To determine whether this was specific to the INF-C enzyme, we analyzed the prototype esterase of group 2 coronaviruses (BCoV HE). A comparison of the relative rates of acetate release catalyzed by INF-C virus and BCoV esterase is shown in Fig. 3c. It is immediately obvious that the rate of acetate release from Neu5,9Ac2α2Me (3) by BCoV HE is higher when compared to the Neu5,9Ac2α(2-6)-S-Glcβ1Me (4). For both enzymes very slow rates of acetate release were detected for Neu5,9Ac2 (2). This result supports the notion that the BCoV HE esterase has a similar mode of action compared to the INF-C virus esterase and engages, most efficiently, only the α-configured 9-O-acetylated substrates. To understand our observed NMR-based assay results at a molecular and atomic level, we re-examined the X-ray crystal structure [24, 30] of INF-C virus HEF in complex with the α-methyl glycoside of 9-acetamido-9-deoxy-N-acetylneuraminic acid (9-acetamido-9-deoxy-Neu5Acα2Me, 6) as shown in Fig. 5. The catalytic triad of the INF-C virus HEF is composed of Ser-57, His-355 and Asp-352. The 9-O-acetate carbonyl oxygen atom orients towards an oxyanion hole formed by the sidechain of asparagine-117 and the NH groups of glycine-85 and Ser-57. The catalytic residue Ser-57 is positioned for nucleophilic attack on the 9-O-acetate carbonyl carbon and the carboxylate group of the N-acetylneuraminic acid moiety forms two hydrogen bonds with arginine (Arg)-322, thus providing enhanced substrate binding. Overall, it is evident that the 9-O-acetate and the carboxylate group are involved in major contacts with the protein. To investigate this further we superimposed 6 with its related β-isomer 9-acetamido-9-deoxy-Neu5Acβ2Me 7. Figure 5 shows the orientation of the carboxylate moiety of 7 assuming an overall identical binding mode. From our molecular modelling studies it is apparent that the esterase could indeed perfectly accommodate a β-methyl glycoside such as 7, but the important bi-dendate interaction of the carboxylate with Arg-322 would be disrupted. We propose that loss of this important interaction provides a rationalisation of why Neu5,9Ac2β2Me (5) is not a suitable substrate for INF-C virus and coronavirus esterases. This result leads us to suggest that there are two absolutely essential pharmacophoric groups in correct spatial arrangement required for strong ligand interaction: the 9-O-acetyl and the C-1 carboxylate groups. We postulate that both moieties are fundamental requirements for the design of a new class of potent esterase inhibitors that are based on carboxylate-containing compounds. Our structural examination of the apo and complexed INF-C virus receptor-destroying esterase domain further revealed some previously unobserved remarkable differences that provide valuable insight for structure-based drug design (Fig. 4). Most striking is the conformational rearrangement of the important catalytic amino acid Ser-57. Attempts to dock (data not shown) Neu5,9Ac2α2Me (3) into the binding site of the apo (blue) INF-C virus esterase structure failed as a result of steric hindrance in the 9-O-acetyl binding domain created by the carbonyl oxygen of Ser-57. It can be seen that the carbonyl oxygen of Ser-57 points into the oxyanion hole of the apo structure (Fig. 5, carbons in brown) preventing the binding of substrate. Upon complexation of 6 the Ser-57 carbonyl group moves down to facilitate an internal hydrogen bond with a His-355-ring nitrogen (Fig. 5, atomic colours). This conformational rearrangement opens the 9-O-acetyl binding site and allows the binding of the O-acetyl moiety. Therefore, the essential amino acid Ser-57 is not only responsible for catalytic activity, but more importantly for binding and orientation of the substrate in the first place.
Fig. 5.
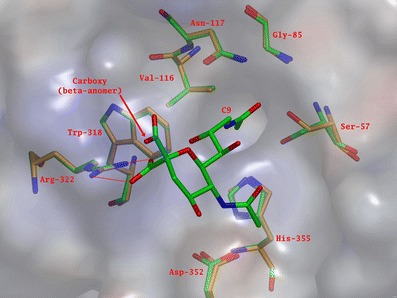
Superimposition of the INF-C virus HEF active site structure. The apo structure carbons are shown in brown and the 9-acetamido-9-deoxy-Neu5Acα2Me (6) INF-C virus HEF complex in atomic colours (αOMe not displayed). The carboxylate group (COO–) of the 9-acetamido-9-deoxy-Neu5Acβ2Me (7) is also shown (cyan). The conformational rearrangement of Ser-57 upon substrate binding is clearly observed
Conclusion
In summary, we have shown that the α-carboxylate and 9-O-acetate moieties are essential pharmacophoric groups. For INF-C virus HEF protein, the rate of acetate release was independent of the type of the aglycon showing identical rates for an O-methyl and an α(2-6)-S-linked glucose moiety. Our examination of the structures of both apo and 6-INF-C virus HEF further suggests that the catalytic amino acid Ser-57 undergoes a conformational change when binding a 9-O-acetylated N-acetylneuraminide. Our combined NMR and molecular modelling study provides new insights into these important enzymes and paves the way for the design and development of novel antiviral therapeutics.
Acknowledgements
The atomic coordinates of the INF-C virus HEF protein-6 complex were kindly provided by Dr. Rosenthal (National Institute for Medical Research, London, UK). The Australian Research Council is acknowledged for its generous support and for the award of a Federation Fellowship (MvI). This work was supported by a short-term fellowship from the European Molecular Biology Organisation ASTF 128.00-06 to JM.
Abbreviations
- INF-C
influenza C
- HEF
haemagglutinin esterase fusion
- BCoV
bovine coronavirus
- HE
haemagglutinin esterase
- NMR
nuclear magnetic resonance spectroscopy
Footnotes
Juliane Mayr and Thomas Haselhorst have made equal contribution.
References
- 1.Matsuzaki Y., Abiko C., Mizuta K., Sugawara K., Takashita E., Muraki Y., Suzuki H., Mikawa M., Shimada S., Sato K., Kuzuya M., Takao S., Wakatsuki K., Itagaki T., Hongo S., Nishimura H. A nationwide epidemic of influenza C virus infection in Japan in 2004. J. Clin. Microbiol. 2007;45:783–788. doi: 10.1128/JCM.01555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevens J., Blixt O., Glaser L., Taubenberger J.K., Palese P., Paulson J.C., Wilson I.A. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Russell R.J., Stevens D.J., Haire L.F., Gamblin S.J., Skehel J.J. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj. J. 2006;23:85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- 4.Rogers G.N., Herrler G., Paulson J.C., Klenk H.D. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J. Biol. Chem. 1986;261:5947–5951. [PubMed] [Google Scholar]
- 5.Herrler G., Klenk H.D. Structure and function of the HEF glycoprotein of influenza C virus. Adv. Virus Res. 1991;40:213–234. doi: 10.1016/S0065-3527(08)60280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrler G., Rott R., Klenk H.D., Muller H.P., Shukla A.K., Schauer R. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. Embo. J. 1985;4:1503–1506. doi: 10.1002/j.1460-2075.1985.tb03809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlasak R., Krystal M., Nacht M., Palese P. The influenza C virus glycoprotein (HE) exhibits receptor-binding (hemagglutinin) and receptor-destroying (esterase) activities. Virology. 1987;160:419–425. doi: 10.1016/0042-6822(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 8.Falk K., Aspehaug V., Vlasak R., Endresen C. Identification and characterization of viral structural proteins of infectious salmon anemia virus. J. Virol. 2004;78:3063–3071. doi: 10.1128/JVI.78.6.3063-3071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellebo A., Vilas U., Falk K., Vlasak R. Infectious salmon anemia virus specifically binds to and hydrolyzes 4-O-acetylated sialic acids. J. Virol. 2004;78:3055–3062. doi: 10.1128/JVI.78.6.3055-3062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smits S.L., Gerwig G.J., van Vliet A.L.W., Lissenberg A., Briza P., Kamerling J.P., Vlasak R., deGroot R.J. Nidovirus sialate-O-acetylesterases: evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J. Biol. Chem. 2005;280:6933–6941. doi: 10.1074/jbc.M409683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlasak R., Luytjes W., Spaan W., Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U S A. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlasak R., Luytjes W., Leider J., Spaan W., Palese P. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J. Virol. 1988;62:4686–4690. doi: 10.1128/jvi.62.12.4686-4690.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultze B., Wahn K., Klenk H.D., Herrler G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology. 1991;180:221–228. doi: 10.1016/0042-6822(91)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klausegger A., Strobl B., Regl G., Kaser A., Luytjes W., Vlasak R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J. Virol. 1999;73:3737–3743. doi: 10.1128/jvi.73.5.3737-3743.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regl G., Kaser A., Iwersen M., Schmid H., Kohla G., Strobl B., Vilas U., Schauer R., Vlasak R. The hemagglutinin-esterase of mouse hepatitis virus strain S is a sialate-4-O-acetylesterase. J. Virol. 1999;73:4721–4727. doi: 10.1128/jvi.73.6.4721-4727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwegmann-Wessels C., Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 2006;23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Groot R.J. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj. J. 2006;23:59–72. doi: 10.1007/s10719-006-5438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vlasak R., Muster T., Lauro A.M., Powers J.C. Palese P, influenza C virus esterase: analysis of catalytic site, inhibition, and possible function. J. Virol. 1989;63:2056–2062. doi: 10.1128/jvi.63.5.2056-2062.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muchmore E.A., Varki A. Selective inactivation of influenza C esterase: a probe for detecting 9-O-acetylated sialic acids. Science. 1987;236:1293–1295. doi: 10.1126/science.3589663. [DOI] [PubMed] [Google Scholar]
- 22.Herrler G., Multhaup G., Beyreuther K., Klenk H.D. Serine 71 of the glycoprotein HEF is located at the active site of the acetylesterase of influenza C virus. Arch. Virol. 1988;102:269–274. doi: 10.1007/BF01310831. [DOI] [PubMed] [Google Scholar]
- 23.Myers M., Richmond R.C., Oakeshott J.G. On the origins of esterases. Mol. Biol. Evol. 1988;5:113–119. doi: 10.1093/oxfordjournals.molbev.a040485. [DOI] [PubMed] [Google Scholar]
- 24.Rosenthal P.B., Zhang X., Formanowski F., Fitz W., Wong C.H., Meier-Ewert H., Skehel J.J., Wiley D.C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strasser P., Unger U., Strobl B., Vilas U., Vlasak R. Recombinant viral sialate-O-acetylesterases. Glycoconj. J. 2004;20:551–561. doi: 10.1023/B:GLYC.0000043292.64358.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mawhinney T.P., Chance L. Hydrolysis of sialic acids and O-acetylated sialic acids with propionic acid. Anal. Biochem. 1994;223:164–167. doi: 10.1006/abio.1994.1564. [DOI] [PubMed] [Google Scholar]
- 28.Haselhorst T., Blanchard H., Frank M., Kraschnefski M.J., Kiefel M.J., Szyczew A.J., Dyason J.C., Fleming F., Holloway G., Coulson B.S., von Itzstein M. STD NMR spectroscopy and molecular modeling investigation of the binding of N-acetylneuraminic acid derivatives to rhesus rotavirus VP8* core. Glycobiology. 2006;17:68–81. doi: 10.1093/glycob/cwl051. [DOI] [PubMed] [Google Scholar]
- 29.Haverkamp J., Schauer R., Wember M., Kamerling J.P., Vliegenthart J.F. Synthesis of 9-O-acetyl- and 4,9-di-O-acetyl derivatives of the methyl ester of N-acetyl-beta-D-neuraminic acid methylglycoside. Their use as models in periodate oxidation studies. Hoppe Seylers Z. Physiol. Chem. 1975;356:1575–1583. doi: 10.1515/bchm2.1975.356.2.1575. [DOI] [PubMed] [Google Scholar]
- 30.Kiefel M.J., Beisner B., Bennett S., Holmes I.D., von Itzstein M. Synthesis and biological evaluation of N-acetylneuraminic acid-based rotavirus inhibitors. J. Med. Chem. 1996;39:1314–1320. doi: 10.1021/jm950611f. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X., Rosenthal P.B., Formanowski F., Fitz W., Wong C.H., Meier-Ewert H., Skehel J.J., Wiley D.C. X-ray crystallographic determination of the structure of the influenza C virus haemagglutinin-esterase-fusion glycoprotein. Acta. Crystallogr. D. Biol. Crystallogr. 1999;55:945–961. doi: 10.1107/S0907444999000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dauber-Osguthorpe P., Roberts V.A., Osguthorpe D.J., Wolff J., Genest M., Hagler A.T. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins. 1998;4:31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]


