Abstract
A miniarray system was developed for the simultaneous detection of porcine circovirus type 1 (PCV1) and type 2 (PCV2) in pigs. The system consists of a polymerase chain reaction (PCR) step to amplify target viral DNA, followed by detection of the amplified DNA using a membrane-anchored probe array and an avidin-alkaline phosphatase (Av-AP) indicator system. The lower limit of detection of PCV using the miniarray was 101.9 tissue culture infectious dose 50 (TCID50)/ml and 102.08TCID50/ml for PCV1 and PCV2, respectively, and 100 viral copies/μl for both PCV1 and PCV2. We validated the miniarray system using 141 lymph node specimens from pigs with suspected postweaning multisystemic wasting syndrome or porcine dermatitis and nephropathy syndrome. Of the 141 samples evaluated, 55 were identified as positive for PCV by the miniarray. Relative to in situ hybridization, the sensitivity and specificity of the miniarray was 100% and 98.9%, respectively. In contrast to other microarray systems, the miniarray does not require a DNA chip reader, since the results can be determined by visual inspection of colorized spots on a nylon membrane. This system represents an effective alternative method for the differential detection of PCV1 and PCV2 in pigs, as well as the maintenance of PCV-free cell lines and pre-screening of commercial vaccines for possible contamination.
Keywords: Miniarray, PCV2, PCV1
Introduction
PCV1 is widespread in the swine population of many countries based on serological surveys (Allan and Ellis 2000), and is a persistent contaminant of the porcine kidney cell line PK-15 (Tischer et al. 1974). Although PCV1 has been recovered from mummified fetuses and one live case of wasting disease (Allan et al. 1995; LeCann et al. 1997), experimental infection of neonates with PCV1 does not result in clinical symptoms of disease. Thus, PVC1 is considered to be a nonpathogenic virus (Allan et al. 1995; Tischer et al. 1986). Recently, three PCV1 isolates were identified in Australia, one from the spleen of a dead weanling pig, a second from the lymph node of an unthrifty weaned pig, and the third from the spinal cord of a newborn pig with associated congenital tremors (Muhling et al. 2006). PCV1 was also detected in 2 of 18 commercial porcine vaccines, despite testing by the manufacturer for the absence of extraneous viruses (Quintana et al. 2006).
In the late 1990's, PCV2 was associated with an emerging disease syndrome in pigs described as postweaning multisystemic wasting syndrome (PMWS) (Allan et al. 1998). Since then, PCV2 has been associated with several other conditions, including congenital tremors, abortion, and porcine dermatitis and nephropathy syndrome (PDNS) (Sorden et al. 1999; Stevenson et al. 2001; West et al. 1999). PCV associated disease (PCVAD) defines a spectrum of symptoms that are associated with PCV2, and of late, this disease has been having a severe impact on swine production worldwide. The differential diagnosis of PCV1 and PCV2 has been reported using virus-specific PCR (Hamel et al. 2000; Larochelle et al. 1999; Liu et al. 2000) and in situ hybridization (Kim and Chae 2001).
We have developed a miniarray system that enables the visual detection of PCV1 and PCV2. Detection is based on a colorimetric reaction following the hybridization of amplified viral DNA that contains a biotinylated nucleotide (biotin-16-dUTP) with oligonucleotide probes that are dotted on a nylon membrane (Quere et al. 2002). Recently, similar DNA miniarray chips for the diagnosis of swine and bovine viruses have been reported. Deregt et al (2006) developed an oligonucleotide suspension microarray (Luminex® Microsphere System) for the differential detection of the animal pestiviruses classical swine fever virus (CSFV), bovine viral diarrhea virus types 1 and 2 (BVDV1 and BVDV2), and border disease virus (BDV). Liu et al (2006) reported a microarray chip assay for foot and mouth disease and porcine reproductive and respiratory syndrome viral genes.
The objective of this study was to develop a DNA miniarray for the simultaneous detection of PCV1 and PCV2 in pigs with non-PMWS, PMWS and/or PDNS. The potential applications of the miniarray system also include the maintenance of PCV-free cell lines and pre-screening of commercial vaccines for contamination.
Materials and methods
Field specimens
We obtained 141 lymph node specimens from archived tissue samples that were collected from pigs nationwide during 2005 and 2006 and submitted to the Pathology Laboratory of the National Veterinary Research and Quarantine Service (NVRQS). The pigs ranged in age from 2 to 4 months. All specimens had been examined for PCV using in-situ hybridization (Kim and Chae 2001). Specimens were classified into two groups according to the presence of PCV genomes in lymphoid tissues (based on in situ hybridization): PCV-negative (n = 87) and PCV-positive (n = 54). The latter group was further subdivided into five sub-groups: PCV1 (n = 1), PCV1 & 2 (non-PMWS; n = 3), PCV2 (non-PMWS; n = 26), PCV2 (PMWS; n = 22) and PCV2 (PDNS; n = 2) using the three criteria method for PMWS diagnosis (Segalés et al. 2005) (Table 1). The clinical characterization of pigs in sub-groups PCV1 and non-PMWS included weak or no respiratory symptoms, and slight or non-existent histopathological lesions. The clinical signs of PDNS included skin lesions, anorexia, severe weight loss and depression. Gross skin lesions were typically round or irregularly shaped, red to purple in color and coalesced over the hind-quarters, limbs and abdomen. Pigs also had enlarged kidneys, with pale cortices with multiple red circular hemorrhagic cortical foci measuring approximately 2–4 mm in diameter. PCV1 from porcine kidney cells (ATCC: CCL31) and isolated PCV2 (Accession No: AF520783) were used as positive controls.
Table 1.
The simultaneous detection of porcine circovirus (PCV) types from 141 lymphoid tissue samples by miniarray and in situ hybridization
| Virus or Non-virus | No. Pigs | Miniarray | in situ hybridization | |||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |||||
| PCV1 | PCV2 | PCV1/2 | PCV1 | PCV2 | ||||
| PCV1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| PCV1&2(non-PMWS) | 3 | 3 | 3 | 3 | 0 | 3 | 3 | 0 |
| PCV2 (non-PMWS) | 26 | 0 | 26 | 26 | 0 | 0 | 26 | 0 |
| PCV2 (PMWS) | 22 | 0 | 22 | 22 | 0 | 0 | 22 | 0 |
| PCV2 (PDNS) | 2 | 0 | 2 | 2 | 0 | 0 | 2 | 0 |
| Non-virus | 87 | 0 | 1 | 1 | 86 | 0 | 0 | 0 |
| Total | 141 | 4 | 54 | 55 | 86 | 4 | 53 | 87 |
DNA extraction
PCV DNA was extracted from 200 μl of lymph node tissue homogenate in Eagle's minimum essential medium (E-MEM, Gibco-BRL) using a G-Spin™ DNA Extraction Kit (iNtRON Biotechnology Co, Korea), according to the manufacturer's instruction.
Amplification of target viral sequences
PCV-specific primers for the amplification of the open reading frame 1 (ORF1) gene of PCV2 (Accession No: AF201897) from the extracted DNA specimens were designed using sequences from the GenBank nucleotide database of the National Center for Biotechnology Information (NCBI). The nucleotide sequences for the sense primer (5′-CCGCTGCCACATCGAGAAAG-3′) and anti-sense primer (5′-CCCGCTCACTTTCAA AAGTTCAGC-3′) were specific for conserved regions of the ORF1 gene, such that target DNA from both PCV1 and PCV2 could be amplified using the same set of primers. PCR amplification was carried out in a total reaction volume of 20 μl containing 5 μl of DNA template, 1×PCR buffer (Biobasic), 1.5 mM MgCl2, 0.2 mM dNTPs, 0.3 mM Biotin-16-dUTP (Roche), 1U Taq DNA polymerase (Biobasic), and 10 pmol of each primer. Amplification was performed using the following conditions: denaturation for 5 minutes (min) at 94°C; 35 cycles of 30 seconds (s) at 95°C, 40 s at 65°C, and 50 s at 72°C; and a final step of 5 min at 72°C. Amplified PCR products were verified by electrophoresis on a 1.2% agarose gel.
Generation of membrane-bound PCV probes
Three probes were developed for the probe array: a PCV1-specific probe (PCV1), a PCV2-specific probe (PCV2), and a probe that was specific for both PCV1 and PCV2 (PCV1/2). Probes were designed based on the ORF1 gene sequences of 7 PCV1 strains and 10 PCV2 strains that were deposited in Genbank (Fig. 1). The sequences of the probes were as follows: PCV1, 5′-GCGGAACCAGGGGAAGT44–3′; PCV2, 5′-AGCGGGAGTCTGGTGACCT42–3′; PCV1/2, 5′-GACCTGTCTACTGCTGTGAGTAC CT36–3′). PCV1, PCV2, and PCV1/2 (10 pmol/μl) were dotted and spread on the right, left, and upper corner, respectively, of a triangular piece of nylon membrane (Hybond-N+, Amersham Biosciences) (Fig. 2B) using the inkjet method and a MATRIX™ 1600 Reagent Dispensing Module (Kinematic Automation Inc., USA). Membranes were then heated for 30 min at 70°C.
Fig. 1.
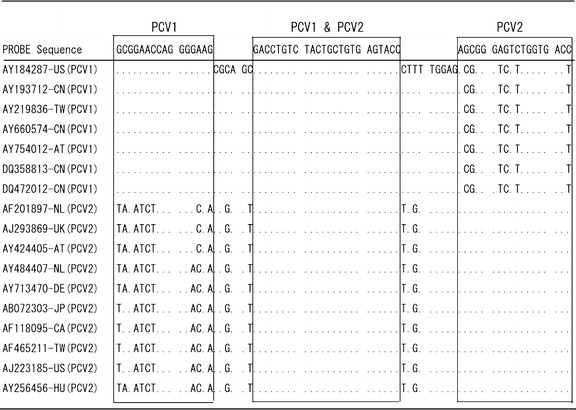
Alignment of the PCV ORF1 gene sequences that were used to design the miniarray probes. The open reading frame (ORF) 1 regions of 7 PCV1 and 10 PCV2 isolates from various countries were used to determine the sequence variation in the probe region. GenBank accession numbers and countries of origin are shown (CA, Canada; CN, China; DE, Germany; AT, Austria; HU, Hungary; JP, Japan; NL, Netherlands; TW, Taiwan; UK, United Kingdom; US, United States of America)
Fig. 2.
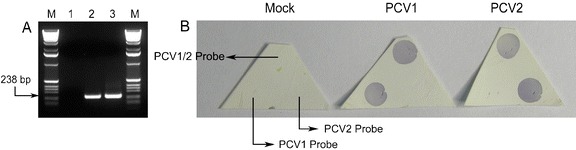
PCR using biotin-16-dUTP and hybridization of PCV1 and PCV2 target sequences. (A) Lane M, 1 kb DNA ladder; Lane 1, mock; Lane 2, PCV1; Lane 3, PCV2. (B) Miniarrays were subjected to hybridization with amplified PCV1 and PCV2 target sequences. Probes were arranged as follows: PCV1/2, upper corner; PCV1 left corner; PCV2, right corner
Hybridization
Membranes containing probes were placed in 1.5 ml microcentrifuge tubes and soaked in prehybridization solution (5×SSC (Roche), 0.5% SDS, 1% blocking solution (Roche)) for 10 min at 45°C. PCR reaction products (10 μl) were mixed thoroughly with 10 μl of denaturation solution (0.5 M NaOH, 1.5 M NaCl) in a microcentrifuge tube and then allowed to stand for 10 min. The prehybridized membrane was transferred to the denatured PCR product mixture, and then incubated in 1 ml of hybridization solution (5×SSC, 0.5% SDS) for 60 min at 45°C. After hybridization, the membrane was washed in 1 ml of washing buffer II (1×SSC, 0.5% SDS) for 5 min at room temperature (RT), and then again for 10 min at 45°C. A third wash was performed in 1 ml of washing buffer I (1×PBS, 0.3% Tween 20 (v/v)) for 3 min at 45°C. For colorimetric detection, membranes were incubated in 1 ml of blocking solution (Roche) for 2 min at RT. Membranes were then incubated with 3 μl of Av-AP (Promega) in 1 ml of blocking solution for 30 min at RT, and then washed twice in 1 ml of washing buffer I for 5 min at RT. The substrate reaction solution was composed of 20 μl of nitrobluetetrazolium and bromochloroindolyl phosphate (NBT/BCIP, Roche) in 1 ml of detection buffer (0.1 M Tris-HCl, 0.1 M NaCl, 0.05 M MgCl, pH 9.5 (20°C)). The reaction was allowed to proceed for 5 to 30 min, until color was visually detected, and then the reaction was stopped by the addition of stop solution (10 mM Tris-HCl, 1 mM EDTA, pH 8.0).
Optimization of assay conditions
To optimize the concentration of the PCV1, PCV2 and PCV1/2 probes, two-fold serial dilutions of each probe (20, 10 and 5 pmol/μl) were hybridized with DNA extracted from PCV1-positive (102.5TCID50/ml) and PCV2-positive (103.0TCID50/ml) controls. To determine the detection limit of Av-AP, PCV DNA was amplified by PCR using biotin-16-dUTP, and then quantified using a spectrophotometer (NanoDrop ND-1000, Nanodrop technologies). Ten-fold serial dilutions of PCV DNA (100, 10, and 1 pg/μl) were prepared and then assayed.
The detection limit of the miniarray for PCV
After optimizing the assay conditions, we determined the lower limit of detection of the miniarray using two-fold serial dilutions of PCV1 (102.5TCID50/ml) and PCV2 (103.0TCID50/ml) viral titers. We also determined the minimum number of viral particles that could be detected by the miniarray. The M r of the 238 basepair (bp) amplified PCV target sequence was estimated to be 1.5708 × 105. We determined the concentration of PCV DNA by absorbance at 260 nm (A260) (NanoDrop ND-1000, Nanodrop technologies), and viral copy number was calculated according to the following formula (Mackay 1999; Tan and Tannock 2005):
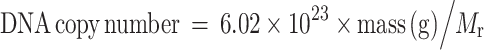 |
Ten-fold serial dilutions (10000, 1000, 100, 10, 1 copies) of the purified 238 bp segment were prepared in parallel as internal standards, amplified, and then subjected to hybridization.
Assay validation using field samples
The miniarray was evaluated for sensitivity and specificity using 141 field specimens that were previously assayed by in situ hybridization. The reproducibility of miniarray was confirmed by re-testing all field samples.
Results
Manufacture of the miniarray
The signature region of the PCV ORF1 gene was amplified by PCR, and the products were separated by 1.2% agarose gel electrophoresis. As expected, a 238 bp PCR product was amplified from PCV1 and PCV2 DNA (Fig. 2A). When analyzed by miniarray, amplified PCV1 and PCV2 targets produced a dark-blue colorimetric reaction at the left and right corners of the membrane probe array, respectively. In both cases, a dark-blue colorimetric reaction was present in the upper corner of the membrane probe array, which contained the PCV1/2 probe (Fig. 2B).
Detection limit of the miniarray for PCV
Viral titers of 101.9TCID50/ml (PCV1) and 102.08TCID50/ml (PCV2) were sufficient to generate a colorimetric signal that was detectable by visual inspection (data not shown). The miniarray was also able to detect amplified target sequences from samples that contained as few as 100 viral DNA copies/μl.
Optimization of assay conditions
The PCV1, PCV2, and PCV1/2 probes were serially diluted two-fold to generate a range of concentrations from 20 to 5 pmol/μl, which were then dotted on the nylon membrane. Target sequences that were amplified by PCR from PCV1 (102.5TCID50/ml) and PCV2 (103.0TCID50/ml) positive controls produced colorimetric reactions at the appropriate spots on the membrane for all probe dilutions tested. Therefore, we selected a concentration of 10 pmol probe for each miniarray (data not shown). The limit of detection of Av-AP was approximately 1 pg DNA (data not shown). The optimum reaction time to obtain a reading with no background was approximately 10 min.
Validation of the miniarray using field samples
Miniarrays consisting of triangular-shaped membrane-bound probe arrays (10 pmol of PCV1, PCV2, and PCV1/2) were used to examine the presence of PCV in 141 lymph node specimens. Of the 141 field specimens examined, 55 clearly showed a PCV-positive reaction. One of the 55 PCV-positive specimens were positive for PCV1, and three specimens were positive for PCV1 & 2 and the remaining 51 were positive for PCV2 (Table 1). Relative to in situ hybridization, the sensitivity and specificity of the miniarray was 100% and 98.9%, respectively. Reproducibility test conducted by re-testing all field samples showed 100% match between the test results.
Discussion
Using a novel miniarray detection system, we were able to successfully detect PCV1- PCV2-target DNA sequences in field samples from pigs. During optimization of the assay, binding to the PCV1-specific probe was consistently negative when biotin-labeled PCV2 DNA (Accession No: AF520783) was applied to the miniarray, which indicated that there is no cross-reactivity between probes. The PCV miniarray primers and probes were designed to specifically amplify PCV1 and PCV2 sequences, and did not match the gene sequences of other porcine pathogens when we searched available sequences using NCBI blast. Thus, the generation of false-positive or false-negative results due to non-specific hybridization is minimized in this assay.
The sensitivity of miniarrays has been shown to be dependent on the concentration of membrane-bound probe (Bertucci et al. 1999). Thus, we required a solid support that had a high capacity to bind small quantities of DNA. The chemical properties of the nylon membrane made it an ideal support for the probe array, because, unlike thin-surface deposition, which is typically used in DNA chip technology, wherein the binding of probes and hybridization are performed on glass or silicon in a three-dimensional system, oligonucleotides can readily penetrate into the nylon membrane of the current miniarray system. Recently, the level of detection of virus by DNA arrays was reported in units of TCID50/ml (viral titer), and copy number (viral particles) (Deregt et al. 2006; Liu et al. 2007). In our study, the sensitivity of the miniarray for PCV1 and PCV2 was similar when measured by TCID50/ml. The minimum viral copy number that could be detected was also similar for the two viruses, and similar to that of the severe acute respiratory syndrome (SARS) virus (de Souza Luna et al. 2007). Interestingly, the concentration of PCV2 in pigs with PMWS and PDNS was higher than in pigs with non-PMWS. At the time of reporting, the non-PMWS pigs exhibited some clinical signs or mild degrees of lesions, but only low levels of PCV2 genome in lymphoid tissue (Fort et al. 2007).
The colorimetric detection system of the current miniarray eliminates the need for specialized equipment, because signals can be detected with the naked eye. However, there is the risk of false-positive or false-negative results due to background interference, such as the false-positive shown for one PCV2-negative sample in this study. When detection is based on a visual reading, as is the case with a variety of commercial antigen or antibody detection kits using immunochromatography method (Anigen Rapid kit, Animal Genetics, Inc), it is important to define the optimal reaction time and virus concentration so as to reduce background color. The optimum reaction time of the current miniarray, during which there was little or no non-specific binding, was approximately 10 min using optimum washing and hybridization.
In situ hybridization is a valuable adjunct to standard DNA extraction techniques for evaluating gene expression in tissues and cells. Its major advantage is the ability to determine which tissues or cells in a mixed population are expressing the DNA of interest. This technique has been used as a standard assay for PCV detection in pigs, and was used to validate the results of the miniarray system. The results of in situ hybridization of the field samples were identical to the results of the miniarray for the detection of PCV1 and PCV2.
In conclusion, we have developed a DNA miniarray for simultaneous detection of PCVs that provides similar level of sensitivity (100%) and specificity (98.9%) as in situ hybridization. This assay can be used to monitor PCV-free cell lines for research and to prescreen commercial vaccines for PCV contamination. This system represents an effective alternative method for the detection and differentiation of PCV1 and PCV2 in pigs.
Acknowledgements
This work was supported partly by Technology Development Program for Agriculture and Forestry, Ministry of Agriculture and Forestry, Republic of Korea.
Abbreviations
- PCV1
porcine circovirus type 1
- PCV2
porcine circovirus type 2
- PMWS
postweaning multisystemic wasting syndrome
- PDNS
porcine dermatitis and nephropathy syndrome
- PCVAD
porcine circovirus associated disease
References
- Allan G.M., McNeilly F., Cassidy J.P., Reilly G.A., Adair B., Ellis W.A., McNulty M.S. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Veterinary Microbiology. 1995;44:49–64. doi: 10.1016/0378-1135(94)00136-K. [DOI] [PubMed] [Google Scholar]
- Allan G.M., McNeilly F., Kennedy S., Daft B., Clarke E.G., Ellis J.A., Haines D.M., Meehan B.M., Adair B.M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. Journal of Veterinary Diagnostic Investigation. 1998;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- Allan G.M., Ellis J.A. Porcine circoviruses: a review. Journal of Veterinary Diagnostic Investigation. 2000;12:3–14. doi: 10.1177/104063870001200102. [DOI] [PubMed] [Google Scholar]
- Bertucci F., Bernard K., Loriod B., Chang Y.C., Granjeaud S., Birnbaum D., Nguyen C., Peck K., Jordan B.R. Sensitivity issues in DNA array-based expression measurements and performance of nylon microarrays for small samples. Human Molecular Genetics. 1999;8:1715–1722. doi: 10.1093/hmg/8.9.1715. [DOI] [PubMed] [Google Scholar]
- Deregt D., Gilbert S.A., Dudas S., Pasick J., Baxi S., Burton K.M., Baxi M.K. A multiplex DNA suspension microarray for simultaneous detection and differentiation of classical swine fever virus and other pestiviruses. Journal of Virological Methods. 2006;136:17–23. doi: 10.1016/j.jviromet.2006.03.025. [DOI] [PubMed] [Google Scholar]
- de Souza Luna L.K., Heiser V., Regamey N., Panning M., Drexler J.F., Mulangu S., Poon L., Baumgarte S., Haijema B.J., Kaiser L., Drosten C. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J Clin Microbiol. 2007;45:1049–52. doi: 10.1128/JCM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort M., Olvera A., Sibila M., Segalés J., Mateu E. Detection of neutralizing antibodies in postweaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected pigs. Vet Microbiol. 2007;125:244–55. doi: 10.1016/j.vetmic.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Hamel A.L., Lin L.L., Sachvie C., Grudeski E., Nayar G.P. PCR detection and characterization of type-2 porcine circovirus. Canadian Journal of Veterinary Research. 2000;64:44–52. [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chae C. Differentiation of porcine circovirus 1 and 2 in formalin-fixed, paraffin-wax-embedded tissues from pigs with postweaning multisystemic wasting syndrome by in situ hybridization. Research in Veterinary Science. 2001;70:265–269. doi: 10.1053/rvsc.2001.0471. [DOI] [PubMed] [Google Scholar]
- Larochelle R., Anataya M., Morin M., Magar R. Typing of porcine circovirus in clinical specimens by multiplex PCR. Journal of Virological Methods. 1999;80:69–75. doi: 10.1016/S0166-0934(99)00032-4. [DOI] [PubMed] [Google Scholar]
- LeCann P., Albina E., Madec F., Cariolet R., Jesin A. Piglet wasting disease. Veterinary Record. 1997;141:660. [PubMed] [Google Scholar]
- Liu Q., Wang L., Willson P., Babiuk L.A. Quantitative, competitive PCR analysis of porcine circovirus DNA in serum from pigs with postweaning multisystemic wasting syndrome. Journal of Clinical Microbiology. 2000;38:3474–3477. doi: 10.1128/jcm.38.9.3474-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Bai Y., Ge Q., Zhou S., Wen T., Lu Z. Microarray-in-a-tube for detection of multiple viruses. Clinical Chemistry. 2007;53:188–194. doi: 10.1373/clinchem.2006.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.C., Huang G.S., Wu M.C., Hong M.Y., Hsiung K.P. Detection of foot and mouth disease and porcine reproductive and respiratory syndrome viral genes using microarray chip. Veterinary Research Communications. 2006;30:191–204. doi: 10.1007/s11259-006-3193-8. [DOI] [PubMed] [Google Scholar]
- Mackay I. Development and evaluation of a quantitative PCR for the determination of cytomegalovirus load. In Abstracts of the XIth International Congress of Virology Sydney Australia VP65. 1999;23:63. [Google Scholar]
- Muhling J., Raye W.S., Buddle J.R., Wilcox G.E. Genetic characterization of Australian strains of porcine circovirus type 1 and 2. Australian Veterinary Journal. 2006;84:421–425. doi: 10.1111/j.1751-0813.2006.00081.x. [DOI] [PubMed] [Google Scholar]
- Quintana J., Segalés J., Calsaminglia M., Domingo M. Detection of porcine circovirus type 1 in commercial pig vaccines using polymerase chain reaction. Veterinary Journal. 2006;171:570–573. doi: 10.1016/j.tvjl.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Quere R., Commes T., Marti J., Bonami J.R., Piquenmal D. White spot syndrome virus and infectious hypodermal and hematopoietic necrosis virus simultaneous diagnosis by miniarray system with colorimetry detection. Journal of Virological Methods. 2002;105:189–196. doi: 10.1016/S0166-0934(02)00106-4. [DOI] [PubMed] [Google Scholar]
- Segalés J., Allan G.M., Domingo M. Porcine circovirus diseases. Anim. Health Res. Rev. 2005;6:119–142. doi: 10.1079/AHR2005106. [DOI] [PubMed] [Google Scholar]
- Sorden S.D., Harms P.A., Nawagitgul P., Cavanaugh D., Paul P.S. Development of a polyclonal-antibody-based immunohistochemical method for the detection of type 2 porcine circovirus in formalin-fixed, paraffin-embedded tissue. Journal of Veterinary Diagnostic Investigation. 1999;11:528–530. doi: 10.1177/104063879901100607. [DOI] [PubMed] [Google Scholar]
- Stevenson G.W., Kiupel M., Mittal S.K., Choi J., Latimer K.S., Kanitz C.L. Tissue distribution and genetic typing of porcine circoviruses in pigs with naturally occurring congenital tremors. Journal of Veterinary Diagnostic Investigation. 2001;13:57–62. doi: 10.1177/104063870101300111. [DOI] [PubMed] [Google Scholar]
- Tan J., Tannock G.A. Role of viral load in the pathogenesis of chicken anemia virus. Journal of General Virology. 2005;86:1327–1333. doi: 10.1099/vir.0.80599-0. [DOI] [PubMed] [Google Scholar]
- Tischer I., Rasch R., Tochtermann G. Characterization of papovavirus- and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol. 1974;226:153–167. [PubMed] [Google Scholar]
- Tischer I., Mields W., Wolff D., Vagt M., Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. Archives of Virology. 1986;91:271–276. doi: 10.1007/BF01314286. [DOI] [PubMed] [Google Scholar]
- West K.H., Bystrom J.M., Wojnarowicz C., Shantz N., Jacobson M., Allan G.M., Haines D.M., Clark E.G., Krakowka S., McNeilly F., Konoby C., Martin K., Ellis J.A. Myocarditis and abortion associated with intrauterine infection of sows with porcine circovirus 2. Journal of Veterinary Diagnostic Investigation. 1999;11:530–532. doi: 10.1177/104063879901100608. [DOI] [PubMed] [Google Scholar]


