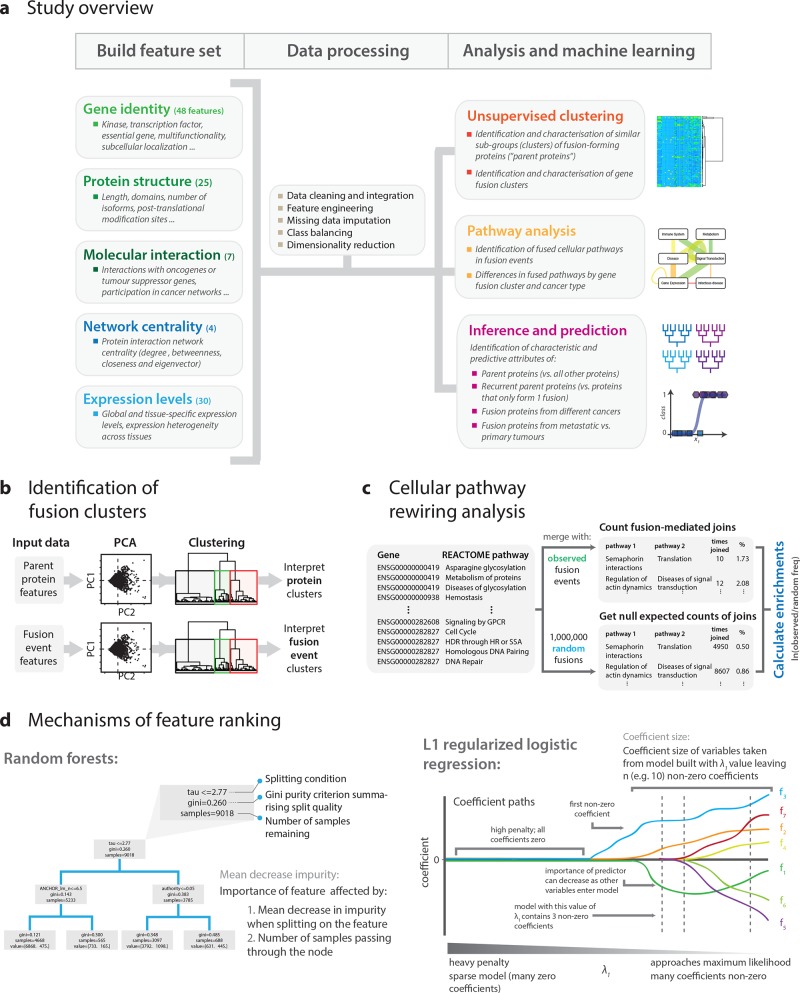
Figure 1.
Study overview. (a) Data sets describing the function, structure, interactions, and expression of human proteins were integrated with a gene fusion data set in order to identify the molecular signatures, hallmarks, and investigate the functional molecular biology of fusion events in cancer. (b) Clusters of fusion-forming proteins (i.e., “parent proteins”) and fusion proteins (each composed of two parent proteins) were identified by principal components analysis followed by agglomerative hierarchical clustering. (c) Cellular pathways significantly rewired by fusion events were identified using randomization tests that compared pathway fusion frequencies to expected null counts. (d) Random forest (RF) and regularized logistic regression (RLR) models were used to infer feature importance across a variety of classification tasks, such as ranking which properties best distinguish between parent proteins and nonfusion forming proteins. The mechanisms of feature importance ranking by the two models are outlined (see Online Methods in the Supporting Information for details).