Abstract
Protease-activated receptor 2 (PAR2) is a G-protein-coupled receptor that is activated by proteolytic cleavage of its N-terminus. The unmasked N-terminal peptide then binds to the transmembrane bundle, leading to activation of intracellular signaling pathways associated with inflammation and cancer. Recently determined crystal structures have revealed binding sites of PAR2 antagonists, but the binding mode of the peptide agonist remains unknown. In order to generate a model of PAR2 in complex with peptide SLIGKV, corresponding to the trypsin-exposed tethered ligand, the orthosteric binding site was probed by iterative combinations of receptor mutagenesis, agonist ligand modifications, and data-driven structural modeling. Flexible-receptor docking identified a conserved binding mode for agonists related to the endogenous ligand that was consistent with the experimental data and allowed synthesis of a novel peptide (1-benzyl-1H[1,2,3]triazole-4-yl-LIGKV) with functional potency higher than that of SLIGKV. The final model may be used to understand the structural basis of PAR2 activation and in virtual screens to identify novel agonists and competitive antagonists. The combined experimental and computational approach to characterize agonist binding to PAR2 can be extended to study the many other G protein-coupled receptors that recognize peptides or proteins.
Keywords: activation, GPCR, molecular docking, orthosteric, PAR2, structure-based drug design
Introduction
Protease-activated receptor 2 (PAR2), a G-protein-coupled receptor (GPCR), is one of four members of the protease-activated receptor family (PAR1–4). PARs have a unique activation mechanism where proteolytic cleavage of the N-terminus is required to reveal a tethered ligand that binds intramolecularly and triggers receptor signaling.1 Endogenous activation of PAR2 by the protease trypsin2 exposes the tethered peptide S36LIGKV–, which activates several different G protein-mediated signaling pathways including calcium mobilization. The PAR1 subtype is also activated by trypsin to reveal a distinct tethered ligand S42FLLRN– and has an overall 37% sequence identity with PAR2.3,4 PARs are involved in a wide range of physiological processes and have been implicated in a number of diseases. However, the only drug currently targeting the protease-activated GPCRs is the PAR1 antagonist vorapaxar, which was approved for prevention of thrombosis.5 The PAR2 subtype has been shown to play roles in pain, migraine,6,7 cancer,8,9 inflammation,10,11 and obesity,12 as well as metabolic and cardiovascular diseases.13,14 For these reasons, PAR2 has attracted great interest as a therapeutic target, but drug development has been challenging due to the unusual activation mechanism and difficulties in identifying small-molecule ligands.
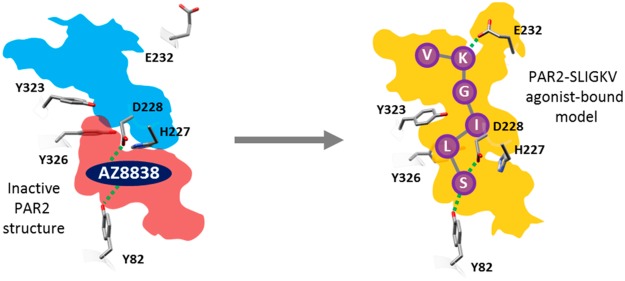
A major breakthrough for structure-based drug design was the recent determination of atomic resolution crystal structures for two PAR subtypes. In 2012, a human PAR1 structure was obtained,15 which revealed the binding mode of the antagonist vorapaxar. More recently, crystal structures of human PAR2 in complex with two different antagonist ligands (AZ8838 and AZ3451) were determined.16 The three PAR antagonists, cocrystallized with their corresponding receptors, bind in distinct pockets. AZ3451 is a noncompetitive antagonist of PAR2 and acts as a negative allosteric modulator. It occupies a site that is formed by transmembrane (TM) helices 2, 3, and 4 and faces the lipid bilayer, whereas the antagonists vorapaxar (PAR1) and AZ8838 (PAR2) bind in two different pockets within the TM bundle of their receptors. Despite these advances in structure determination for PARs, the mechanism of activation by the tethered peptide agonists remains unclear. The synthetic hexapeptide corresponding to the last six amino acids of the human PAR2 tethered ligand, SLIGKV-NH2, is itself an agonist,17 indicating that interactions with the cleaved N-terminus are required for receptor activation. Understanding how peptides and mimetics thereof (e.g., 2-furoyl(2f)-LIGRL-NH2,18 2f-LIGRLO-NH2,19 and GB110)20,21 bind to human PAR2 would facilitate the design of small-molecule ligands which could be developed into future therapeutics. However, as reflected by a community-wide assessment of GPCR structure prediction, modeling of peptide binding is very challenging due to the size and flexibility of such ligands and their interactions with the highly variable loop regions.22 Predictions of agonist recognition by PAR2 have relied on molecular docking to homology models, which has resulted in several different potential binding modes.23,24 Even though atomic resolution structures of PAR2 are now available, modeling of the agonist-bound state is limited by them being determined in an inactive conformation and by the identified binding pockets being too small to accommodate any of the available peptide agonists.
Herein, we combined site-directed mutagenesis of human PAR2 with chemical variation of the peptide ligands and computational modeling to predict the binding mode of agonist peptide SLIGKV. In combination with functional assays, this approach identified the orthosteric site and key interactions of SLIGKV. Molecular docking calculations were used to generate a model of SLIGKV bound to PAR2 consistent with the experimental data. Models obtained via a ligand-based approach that exploited similarities between synthetic PAR2 ligands and AZ8838 further corroborated the proposed binding mode. Our findings highlight interactions of the human PAR2 receptor that are important for agonist and antagonist design which was used to guide discovery of a novel, highly potent, and selective PAR2 agonist.
Results and Discussion
Mapping of PAR2−SLIGKV Interactions by Mutagenesis
To map interactions between the N-terminal region S36LIGKV– and PAR2, we employed an extensive, combinatorial experimental strategy that introduced point mutations in both receptor and agonist. Previous studies have focused exclusively on mutagenesis of the TM regions of the receptor,24 mutagenesis of the tethered ligand,25 mutagenesis of the peptide,26,27 or extracellular loop 2 (ECL2).28,29 In this work, a total of 24 different point mutations were made in PAR2, and 26 different peptides were evaluated in a calcium mobilization functional assay. The screened receptor mutants were selected based on the PAR2 crystal structure and covered the occluded binding site of AZ8838 as well as the neighboring pocket facing the extracellular region. Complementary modifications to the peptide agonist were also designed (Figure S1). All the tested peptides were amidated at the C-terminus. The receptor mutations did not affect receptor expression, and only peptides selective for PAR2 were considered (Figure S2). The 1321N1 cell line used in this study has endogenous levels of PAR1 expression. Therefore, the presented lack of activity of these peptides shows that there are no off-target effects, including no PAR1 activity. Competition binding assays using [3H]-GB110 and a selection of peptides from this study were performed on wild type (WT) receptors (Figure S3 and Table S1). In these cases, peptide binding affinity had the same rank order as potency, consistent with previous results using a [3H]propionyl-2f-LIGRLO-NH2 probe and unlabeled SLIGKV-NH2, SLIGRL-NH2 (the rat sequence), and 2f-LIGRLO-NH2 peptides.19 However, determination of binding affinity at mutant PAR2 is challenging due to the lack of a high-affinity endogenous agonist that can be labeled. In a recent study,24 eight PAR2 mutants were tested against 2f-LIGRLO(dtpa)-NH2, and although saturation binding affinity was comparable with calcium assay potency, the most functionally significant could not be fully probed as high enough concentrations of labeled ligand could not be reached. In the absence of a high affinity endogenous ligand mimetic, this mutagenesis study of the PAR2 receptor, like others before it,24,26,30 was performed by monitoring calcium signaling. It should be noted that a limitation of this approach is that functional potency does not always correlate with affinity as mutants could alter receptor dynamics in a way that influences interactions with intracellular effectors without affecting binding. For example, mutations had much smaller effects on binding affinity compared to potency in a recent study of PAR1.31
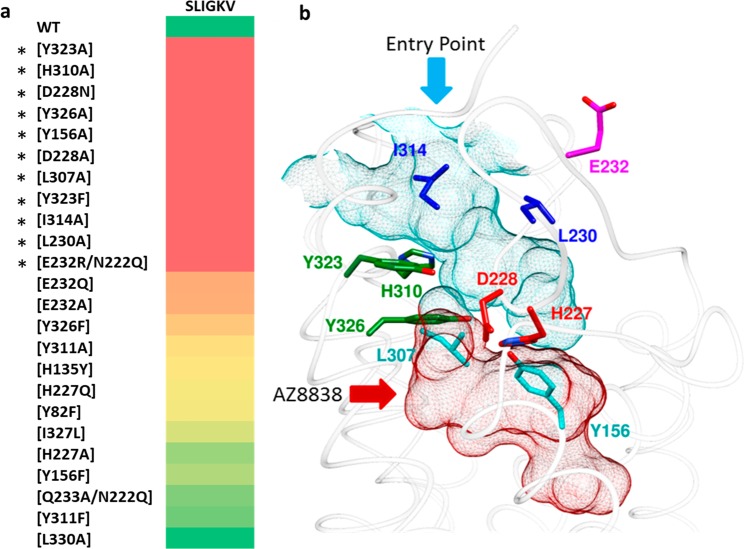
The functional calcium potency readout allowed identification of specific receptor-agonist interactions necessary for activation. Nine receptor mutations led to a >10-fold reduction in potency of SLIGKV compared to the effect at WT PAR2 (Table 1 and Figure 1a). SLIGKV produced no response at the highest concentrations tested for the D228NECL2, H310A6.58, and Y323A7.32 mutants (superscripts represent generic residue numbering system for GPCRs),32 corresponding to a >342-fold drop in potency. Although synthetic peptides SLIGKV-NH2, SLIGRL-NH2, and mimetics thereof are extensively used to improve understanding of the activation mechanism of the PAR2 receptor,8,17−19 the endogenous agonist trypsin was also considered to confirm the relevance of our findings. WT and mutant PAR2 receptors were screened for trypsin-induced activation (Figure S4 and Table S2) and identified that modification of the same receptor residues (Y1563.33, D228ECL2, H3106.58, and Y3237.32) had the largest effect irrespective of protease- or peptide-induced activation of PAR2. The residues that upon mutation led to the largest changes in activity were hypothesized to form key interactions with the peptide and provided a starting point for experiment-guided structure-based modeling of its binding mode (Figure 1b).
Table 1. Potency Data of SLIGKV Peptide at WT and Mutant PAR2 Receptorsa.
| pEC50 | fold change | |
|---|---|---|
| WT | 6.06 ± 0.03 | 1 |
| Y323A7.32 | <3.5 | >342 |
| H310A6.58 | <3.5 | >342 |
| D228NECL2 | <3.5 | >342 |
| Y326A7.35 | 3.69 ± 0.04 | 234 |
| Y156A3.33 | 3.93 ± 0.06 | 135 |
| D228AECL2 | 3.96 ± 0.04 | 124 |
| L307A6.55 | 4.00 ± 0.06 | 113 |
| Y323F7.32 | 4.36 ± 0.09 | 50 |
| I314A6.62 | 4.7 ± 0.1 | 23 |
| L230AECL2 | 4.82 ± 0.05 | 17 |
| E232RECL2/N222QECL2 | 4.85 ± 0.03 | 16 |
| E232AECL2 | 5.18 ± 0.04 | 7 |
| E232QECL2 | 5.18 ± 0.07 | 8 |
| Y326F7.35 | 5.25 ± 0.03 | 6 |
| Y311A6.59 | 5.31 ± 0.03 | 6 |
| H135Y2.64 | 5.33 ± 0.05 | 5 |
| Y82F1.39 | 5.39 ± 0.03 | 5 |
| H227QECL2 | 5.39 ± 0.04 | 5 |
| I327L7.36 | 5.45 ± 0.06 | 4 |
| Y156F3.33 | 5.54 ± 0.05 | 3 |
| H227AECL2 | 5.60 ± 0.05 | 3 |
| Q233AECL2/N222QECL2 | 5.67 ± 0.03 | 2 |
| Y311F6.59 | 5.72 ± 0.04 | 2 |
| L330A7.39 | 5.97 ± 0.03 | 1 |
pEC50 values, measured by calcium mobilization assays, are shown as mean ± s.e.m. (n = 3 independent experiments), and fold change is calculated compared to response at WT.
Figure 1.
Identification of the orthosteric binding site. (a) Heat plot of fold change of SLIGKV-induced activation of calcium mobilization at mutant PAR2 receptors presented on a color scale where red and green correspond to >10-fold drop in potency and effect similar to that of WT, respectively. Numerical data are presented in Table 1. Asterisks highlight the residues identified as important for SLIGKV-induced activation of PAR2. (b) Crystal structure of PAR2 (PDB ID: 5NDD)16 with side chains of key residues shown as sticks. The predicted orthosteric binding site is depicted as a surface mesh, encompassing an entry point from the extracellular space (cyan) and the AZ8838 binding site (red). Residues have been color coordinated based on their spatial arrangement.
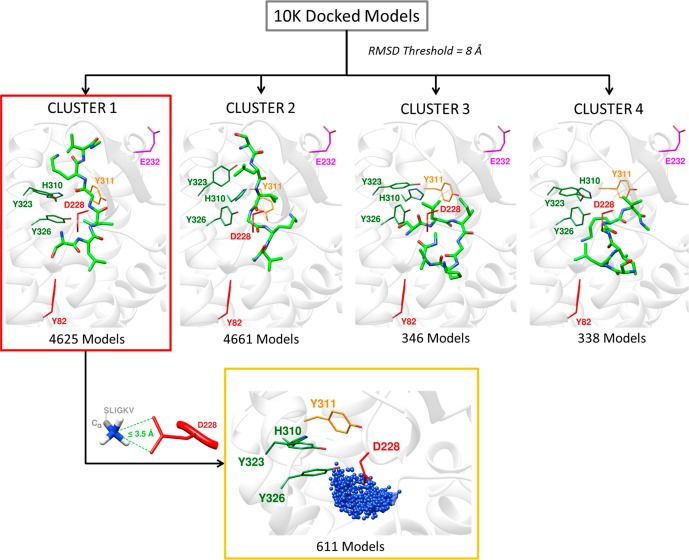
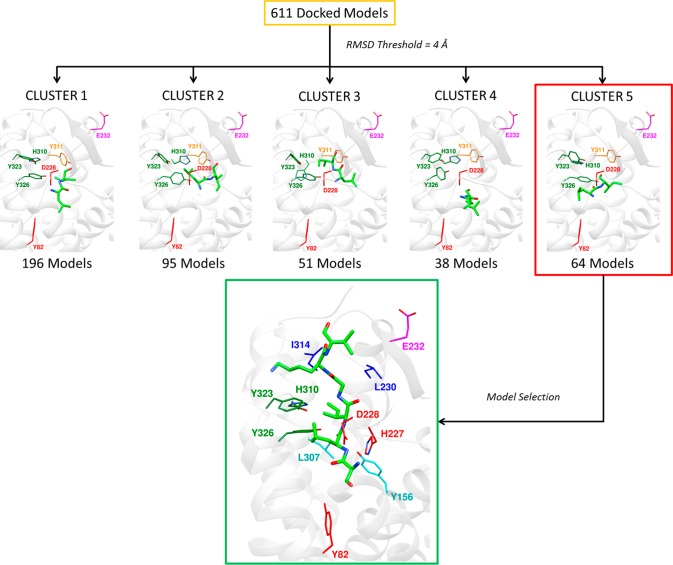
Attempts to model SLIGKV bound to PAR2 using the (rigid) receptor conformation represented by the crystal structure indicated that it was not possible to obtain a binding mode consistent with all the experimental data. The mutagenesis data showed that the peptide agonist interacts with residues in two distinct pockets. This is illustrated in Figure 1b, which depicts the two subpockets of the proposed orthosteric site, comprising an entry point from the extracellular space (cyan) and the occluded AZ8838 binding pocket (red), which is closed off by H227ECL2 and Y3267.35 (Figure S5). Therefore, in order to explore potential peptide binding modes, extensive molecular docking to the PAR2 crystal structure (PDB ID: 5NDD)16 was performed with AutoDock.33 To facilitate accommodation of the agonist, several of the residues identified as important by mutagenesis (H3106.58, Y3237.32, and Y3267.35) were considered flexible during the docking simulations. SLIGKV showed a modest decrease in potency when H227ECL2 and Y3116.59 were mutated to Ala (3- and 6-fold respectively, Table 1), but including flexibility for these side chains allowed the peptide to access both pockets of the proposed orthosteric site. In addition, Q233ECL2 was also considered flexible to enlarge the entrance route for SLIGKV. A total of 10 000 models, each with a unique receptor and ligand conformation, were generated. The resulting binding modes (Figure 2) were analyzed by hierarchical clustering to identify those that agreed with the experimental data. An overview of the structure-based modeling strategy is shown in Figure S6. A first cluster analysis of the peptide conformations was performed by applying an RMSD threshold of 8 Å, and clusters that captured the expected overall binding mode of the peptide were identified. As the SLIGKV peptide is endogenously tethered to the receptor, solutions that did not have the C-terminus facing the extracellular surface were excluded. This criterion reduced the number of models from 10 000 to a single cluster of solutions containing 4625 structures (Figure 2).
Figure 2.
Filtering of PAR2–SLIGKV complexes obtained from molecular docking based on expected placement of N- and C-termini. The 10 000 generated models from molecular docking were clustered, and the best poses from the four largest clusters are shown. Cluster 1 was selected as the position of the C-terminal was facing the extracellular surface, which is compatible with a tethered endogenous peptide. Then, the models from Cluster 1 with a distance >3.5 Å between the serine α-amino group (−NH3+) nitrogen and one of the two carboxylate oxygens of D228ECL2 were discarded. Residues have been color-coordinated based on their spatial arrangement. Blue spheres represent the α-amino group nitrogen of the serine in SLIGKV.
Interactions with D228ECL2 and Y821.39 Anchor SLIGKV in the Binding Site
D228ECL2 is part of the AZ8838 binding pocket (Figure 1b) and forms a hydrogen bond to the imidazole ring of the antagonist in the crystal structure (Figure S5).16 D228ECL2 is conserved in all four PARs and has been reported as a key interacting partner in PAR1 activation.15,31 As it is the only buried, negatively charged residue facing the predicted orthosteric site, we hypothesized that D228ECL2 forms a salt bridge with the N-terminal charge of SLIGKV. The D228AECL2 mutation led to a large decrease in potency (124-fold), suggesting that D228ECL2 is a key interacting partner of SLIGKV (Table 2). Similarly, the D228NECL2 mutation, lacking the side chain charge but maintaining the size of the residue, had a >342-fold loss of activity, supporting the idea that the interaction with the charge is important. The significance of interactions with D228ECL2 was further probed by testing modified peptide N-(3-OH-2-Me-propanoyl)-LIGKV, which lacks the charged N-terminal amine functionality of SLIGKV (Figure S1). Reduction in potency was observed for the combinations of SLIGKV/D228AECL2 (124-fold) and N-(3-OH-2-Me-propanoyl)-LIGKV/WT (36-fold), suggesting that both D228ECL2 and the N-terminal charged moiety are key for activity. On the basis of these results, the ensemble of 4625 models was filtered to exclude the structures that did not have a hydrogen bond between the N-terminal nitrogen and the D228ECL2 side chain carboxylate (>3.5 Å between N and O). This criterion reduced the number of candidate models to 611 (Figure 2). AZ8838 also forms a hydrogen bond to Y821.39 in the crystal structure (Figure S5), which provided a potential candidate to interact with the hydroxyl group of the N-terminal serine. Removing the side chain hydrogen bond donor of the agonist (ALIGKV) led to a 9-fold decrease in potency compared to that of SLIGKV (Table 2) and a similar 5-fold decrease in potency of SLIGKV was observed when the hydroxyl group was removed from Y821.39 (Y82F). If the N-terminal charge of the peptide was removed (N-(3-OH-2-Me-propanoyl)-LIGKV) together with removal of the hydrogen bonding capacity of Y821.39 (Y82F), then an even larger decrease in potency was observed (66-fold), suggesting that loss of multiple polar interactions has an additive effect in this pocket. As the N-terminus cannot interact with both D228ECL2 and Y821.39 based on their position in the crystal structure, these experiments suggested that the serine hydroxyl of SLIGKV forms a hydrogen bond with Y821.39.
Table 2. Potency Data Probing N-Terminal Interactions of Peptide Agonists and PAR2 Receptor with Relevant Single Point Mutationsa.
| SLIGKV | ALIGKV | N-(3-OH-2-Me-propanoyl)-LIGKV | |
|---|---|---|---|
| WT | 6.06 ± 0.03 | 5.10 ± 0.03 | 4.45 ± 0.03 |
| Y82F1.39 | 5.39 ± 0.03 | 4.02 ± 0.08 | 3.57 ± 0.02 |
| D228AECL2 | 3.96 ± 0.04 | <3.5 | <3.5 |
| D228NECL2 | <3.5 | <3.5 | <3.5 |
pEC50 values, measured by calcium mobilization assays, are presented as mean ± s.e.m. (n = 3 independent experiments).
Hydrophobic Side Chains in Positions 2 and 3 of SLIGKV Are Important for Potency
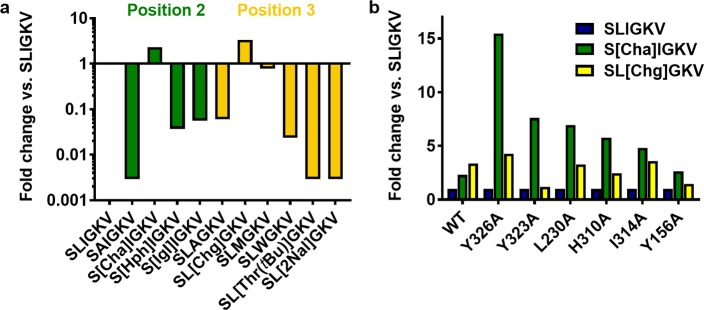
Leucine and isoleucine at positions 2 and 3 of the peptide agonist were modified to assess the size of the pockets available to these residues (Table 3 and Figure 3a). Alanine substitution of the leucine in position 2 of SLIGKV resulted in loss of functional activity at all concentrations tested. In contrast, increasing the size of the side chain to a cyclohexylalanine (Cha) improved potency by 2-fold. Larger variants of the side chain such as homophenylalanine (Hph) and indanylglycine (Igl) resulted in a loss of potency (27- and 18-fold, respectively; Figure 3a). The importance of the leucine residue was further exemplified by alanine substitution of the last four amino acids (SLAAAA), which produced only a 61-fold drop in potency compared to that of SLIGKV (Table 3). Consistent with an extensive study of PAR1 and PAR2 peptide agonists,26 we found that leucine at position 2 of SLIGKV was vital for PAR2 activation and specificity. Some designed peptides (e.g., SFIGKV, SWIGKV, and S[Phe(3,4-diChloro)]IGKV) were excluded as they showed a response in the parental cell line and were suspected of having PAR1 activity. At position 3, a bulky hydrophobic side chain was also found to be favorable, as alanine substitution caused a 17-fold loss of potency. A small increase in side chain size to cyclohexylglycine (Chg) or methionine improved (3-fold) and maintained potency, respectively. However, as in the case of position 2, the pocket size was limited. Tryptophan caused a 43-fold reduction in potency, and 2-naphthylalanine (2Nal) and O-tert-butyl threonine (Thr(tBu)) substitutions led to >342-fold loss of activity (Figure 3a). The data therefore supported that both position 2 and 3 of SLIGKV were buried in the orthosteric site. This result was reproduced by the selected cluster of docking solutions, but among the 611 models, different hydrophobic pockets were explored by the leucine and isoleucine residues.
Table 3. Potency Data Probing Hydrophobic Interactions of Mutant Peptide Agonists and WT PAR2 Receptora.
| peptide | pEC50 ± s.e.m. | peptide | pEC50 ± s.e.m. |
|---|---|---|---|
| SLIGKV | 6.06 ± 0.03 | SLAGKV | 4.83 ± 0.03 |
| SAIGKV | <3.5 | SL[Chg]GKV | 6.58 ± 0.04 |
| S[Cha]IGKV | 6.41 ± 0.05 | SLMGKV | 5.94 ± 0.05 |
| S[Hph]IGKV | 4.61 ± 0.06 | SLWGKV | 4.43 ± 0.07 |
| S[Igl]IGKV | 4.81 ± 0.06 | SL[Thr(tBu)]GKV | <3.5 |
| SLAAAA | 4.27 ± 0.05 | SL[2Nal]GKV | <3.5 |
pEC50 values, measured by calcium mobilization assays, are presented as mean ± s.e.m. (n = 3 independent experiments).
Figure 3.
Interactions of hydrophobic side chains in positions 2 and 3 of SLIGKV. (a) Fold change in potency of modified peptides at WT calculated compared to that of SLIGKV, presented as the log transformed data such that more potent peptides show an increase and less potent peptides show a decrease. (b) Difference in fold change of potency of modified peptides at modified receptors, calculated compared to that of SLIGKV at each receptor. Peptide structures are shown in Figure S1, and potency data are presented in Tables 3 and 4.
To investigate which subpockets of the orthosteric site accommodate positions 2 and 3 of SLIGKV, the modified peptides S[Cha]IGKV and SL[Chg]GKV were screened against PAR2 mutants (Table 4). The fold change in potency of each peptide compared to that of SLIGKV was calculated at each receptor, and the receptor mutations that caused the largest changes are shown in Figure 3b. The most striking results from these experiments were those obtained for the Y326A7.35 mutant. In the case of S[Cha]IGKV, a 2-fold improvement in potency compared to that of SLIGKV was observed at WT, whereas a much larger 15-fold increase was obtained for Y326A7.35. In contrast, SL[Chg]GKV showed similar improvements of potency for WT and the Y326A7.36 mutant. A comparable, but not as pronounced, difference for S[Cha]IGKV was observed for the neighboring residue Y3237.32 (Y323A). These results suggested that the second residue of SLIGKV occupies a pocket formed by Y3237.32 and Y3267.35, in which the larger Cha side chain could be better accommodated if either of these residues were mutated to an alanine. Concentration-response curves of SLIGKV, S[Cha]IGKV, and SL[Chg]GKV at WT and mutants Y323A7.32 and Y326A7.35 are shown in Figure S7. No corresponding strong correlations were identified for SL[Chg]GKV at the screened mutants (Figure 3b). This could be explained by the fact that the pocket for the third residue of SLIGKV is likely to be located closer to the extracellular surface and ECL2 of the receptor and could hence be more flexible than those occupied by the first two residues. The ensemble of 611 remaining models was clustered based on the positions of the leucine and isoleucine residues of SLIGKV using an RMSD threshold of 4 Å (Figure S6). A total of 18 clusters were obtained, and these poses were visually inspected and filtered primarily based on interactions with Y3237.32 and Y3267.35 (Figure 4). The fifth cluster with 64 models placed the leucine in a pocket composed of Y3237.32 and Y3267.35, and the isoleucine was positioned in a pocket in the vicinity of Y1563.33, L230ECL2, and I3146.62, which was consistent with the experimental results for the receptor mutants. A representative model from this cluster is shown in Figure 4.
Table 4. Potency Data Probing Interactions of SLIGKV, S[Cha]IGKV, and SL[Chg]GKV at Mutant PAR2 Receptorsa.
| SLIGKV | S[Cha]IGKV | SL[Chg]GKV | |
|---|---|---|---|
| WT | 6.06 ± 0.03 | 6.41 ± 0.05 | 6.58 ± 0.04 |
| Y323A7.32 | <3.5 | 4.40 ± 0.05 | 3.60 ± 0.04 |
| H310A6.58 | <3.5 | 4.28 ± 0.06 | 3.91 ± 0.06 |
| Y326A7.35 | 3.69 ± 0.04 | 4.87 ± 0.05 | 4.32 ± 0.05 |
| Y156A3.33 | 3.93 ± 0.06 | 4.35 ± 0.05 | 4.09 ± 0.06 |
| L307A6.55 | 4.00 ± 0.06 | 4.72 ± 0.05 | 4.58 ± 0.06 |
| Y323F7.32 | 4.36 ± 0.09 | 5.4 ± 0.1 | 5.23 ± 0.08 |
| I314A6.62 | 4.7 ± 0.1 | 5.38 ± 0.05 | 5.26 ± 0.04 |
| L230AECL2 | 4.82 ± 0.05 | 5.7 ± 0.1 | 5.33 ± 0.06 |
| Y326F7.35 | 5.25 ± 0.03 | 5.94 ± 0.05 | 5.84 ± 0.05 |
| Y311F6.59 | 5.72 ± 0.04 | 6.44 ± 0.05 | 6.49 ± 0.05 |
| Y156F3.33 | 5.54 ± 0.05 | 5.96 ± 0.06 | 6.08 ± 0.06 |
| L330A7.39 | 5.97 ± 0.03 | 6.85 ± 0.06 | 6.53 ± 0.05 |
| Y311A6.59 | 5.31 ± 0.03 | 6.02 ± 0.07 | 6.01 ± 0.05 |
| I327L7.36 | 5.45 ± 0.06 | 6.06 ± 0.05 | 6.15 ± 0.05 |
pEC50 values, measured by calcium mobilization assays, presented as mean ± s.e.m. (n = 3 independent experiments).
Figure 4.
Cluster analysis based on the second and third residues of SLIGKV (leucine and isoleucine, respectively) and model selection. The best poses from the top five clusters are depicted. Receptor residues have been color-coordinated based on their spatial arrangement.
Interaction with E232ECL2 at the Receptor Surface
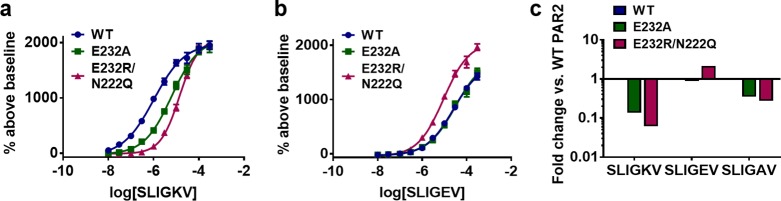
If E232ECL2 in ECL2 was mutated to an alanine (E232AECL2) or if only the charge was removed (E232QECL2), then there was a 7- or 8-fold loss in SLIGKV potency, respectively (Table 5). A larger 16-fold change in SLIGKV potency was observed with charge reversal to a positively charged arginine (E232RECL2/N222QECL2, Table 5, Figure 5a,c). The E232RECL2/N222QECL2 mutant was already available from a prior study, and the single N222QECL2 mutation, located in ECL2, did not have a significant effect on SLIGRL-NH2 potency in calcium signaling assays.16 A charge-reversed peptide (SLIGEV) displayed an activity pattern opposite to that of SLIGKV. SLIGEV had the lowest potency at WT and the neutral mutants (E232AECL2 and E232QECL2), whereas it was most potent at the E232RECL2/N222QECL2 mutant receptor, as salt bridge formation was then possible (Table 5, Figure 5b,c). Removal of the lysine charge in the peptide agonist (SLIGAV) showed smaller effects at the E232ECL2 mutants, and the pattern of regaining potency was not observed with the E232RECL2/N222QECL2 mutant (Table 5, Figure 5c). The mutagenesis data therefore suggested that there is a salt bridge between the lysine of the peptide and E232ECL2. A previous study also highlighted the importance of E232ECL2 in peptide activation of the rat PAR2 receptor.28 Furthermore, a similar charge-reversal experiment for PAR1 reached the same conclusion that SFLLRN-NH2 regained activity on the corresponding (E260RECL2) mutant receptor compared to that of the SFLLEN-NH2 peptide at WT.34 No charge-charge interactions between the lysine of the peptide and E232ECL2 were found among the remaining 64 models of PAR2–SLIGKV. This appeared to be due to the conformation of ECL2, which oriented the side chain away from the entry point of the proposed orthosteric site. To explore if this interaction could be formed, refinement was performed for a representative model using MODELLER,35 which demonstrated that the charge-charge interaction was feasible after only small rearrangements of the receptor binding site, ECL2, and peptide. This resulted in our final model of SLIGKV bound to PAR2 based on molecular docking (Figure 6).
Table 5. Potency Data Probing C-Terminal Interactions of Peptide Agonists at Mutant PAR2 Receptorsa.
| SLIGKV | SLIGEV | SLIGAV | |
|---|---|---|---|
| WT | 6.06 ± 0.03 | 4.69 ± 0.06 | 5.16 ± 0.04 |
| E232AECL2 | 5.18 ± 0.04 | 4.64 ± 0.06 | 4.70 ± 0.08 |
| E232QECL2 | 5.18 ± 0.07 | 4.44 ± 0.07 | 4.64 ± 0.09 |
| E232RECL2/N222QECL2 | 4.85 ± 0.03 | 5.02 ± 0.05 | 4.60 ± 0.05 |
pEC50 values, measured by calcium mobilization assays, are presented as mean ± s.e.m. (n = 3 independent experiments).
Figure 5.
C-terminal interactions between PAR2 and SLIGKV. Concentration-response curves of calcium signaling by (a) SLIGKV and (b) SLIGEV at WT (blue) and mutant receptors E232A (green) and E232R/N222Q (magenta). Data are presented as mean ± s.e.m. of n = 3 independent experiments, and numerical values are shown in Table 5. (c) Fold change in potency calculated compared to peptide response at WT.
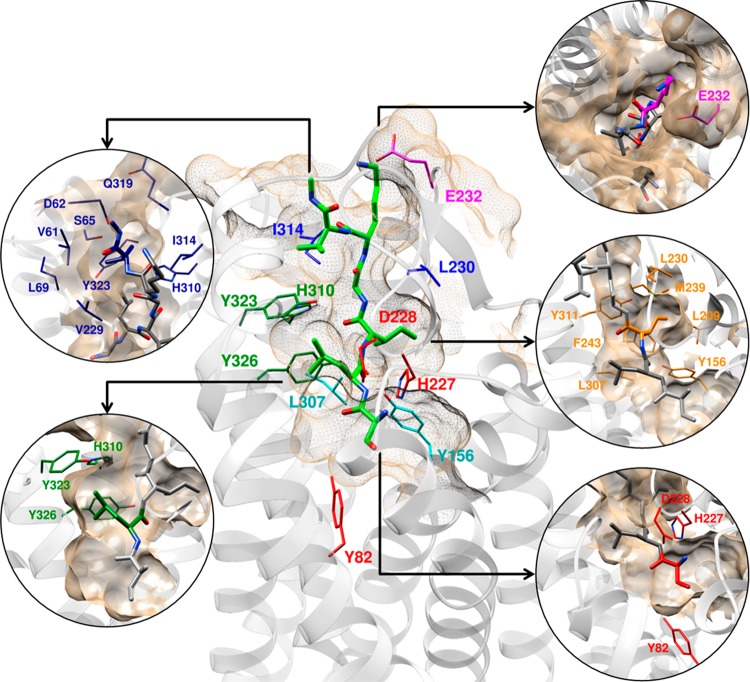
Figure 6.
Refined model of SLIGKV in PAR2 from molecular docking calculations. Peptide residues and key protein side chains are highlighted. Receptor residues have been color-coordinated based on their spatial arrangement.
Analysis of Binding Modes for Synthetic PAR2 Ligands and Design of Potent Novel Agonists
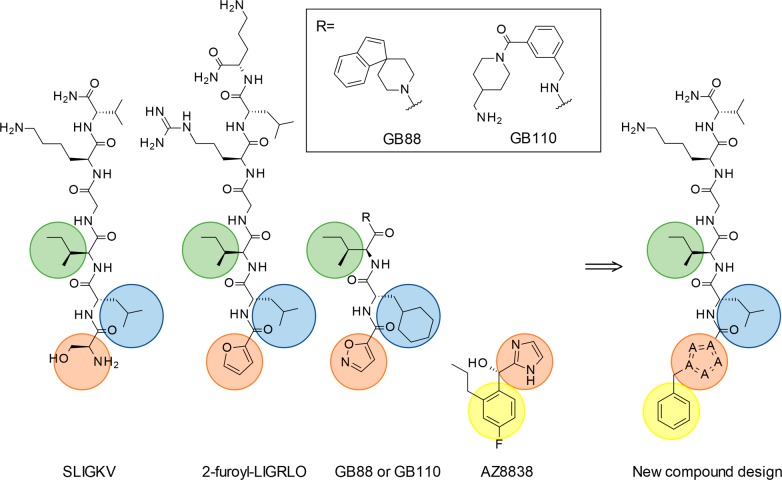
Further validation of our model for agonist binding was carried out by analyzing synthetic PAR2 ligands. In parallel with the molecular docking study, a ligand-centric approach was employed to compare the binding modes of SLIGKV to a series of other agonists (Figure 7). The key pharmacophoric groups of SLIGKV and previously reported PAR2 ligands 2f-LIGRLO, GB110, and GB88 were assumed to have overlapping binding modes. This placed the heterocyclic groups of the synthetic ligands in the same position as the N-terminus of SLIGKV. On the basis of the mutagenesis experiments, which indicated that AZ8838 mimics the polar interactions made by the N-terminus of the endogenous peptide, the imidazole group of AZ8838 would overlap with the heterocyclic groups of 2f-LIGRLO, GB88, and GB110 (Figure 7). To construct a shape-based overlay with SLIGKV using this ligand-based approach, a conformational search for GB88 was performed as it had the lowest number of rotatable bonds among the ligands, which reduced the number of poses to consider. The resulting ensemble was filtered using the program ROCS36 to identify conformations that matched the shape of AZ8838 and carbon atoms placed in positions hypothesized to be occupied by positions 2 and 3 of SLIGKV in the binding site. An overlay of the best fitting pose of GB88 to the PAR2 crystal structure (PDB ID: 5NDD)16 displayed clashes with Y3116.59, Y3237.32, and Y3267.35. Optimization of the complex with GB88 placed in the binding site using the program Prime37 demonstrated that the predicted conformation could be accommodated after rearrangements of a number of protein side chains. Y3116.59, Y3237.32, and Y3267.35 showed a heavy atom RMSD of 2.2, 4.2, and 1.7 Å, between the experimental and optimized receptor structures, respectively. In agreement with the molecular docking results, this second model suggested significant side chain rearrangements for Y3116.59, Y3237.32, and Y3267.35 between the AZ8838- and agonist-bound complexes. The first four residues of SLIGKV and 2f-LIGRLO could also be placed in this model without clashes by using a shape-based alignment to the predicted binding conformation of GB88. The heterocycles of GB88 and 2f-LIGRLO overlapped with both the imidazole group of AZ8838 and the N-terminal part of SLIGKV in our model.
Figure 7.
PAR2 ligands SLIGKV, 2f-LIGRLO, GB110, and GB88 display similar pharmacophoric groups with a polar or heterocyclic motif in the first position (orange) and a hydrophobic residue in the second (blue) and third (green) positions. The models of SLIGKV and GB88 suggested that the first position was overlapping with the imidazole moiety of the small molecule antagonist AZ8838 and that none of the agonists exploited the pocket occupied by the 4-fluoro-2-propylphenyl moiety. New compounds that probed a lipophilic extension of the agonist SLIGKV were designed (yellow).
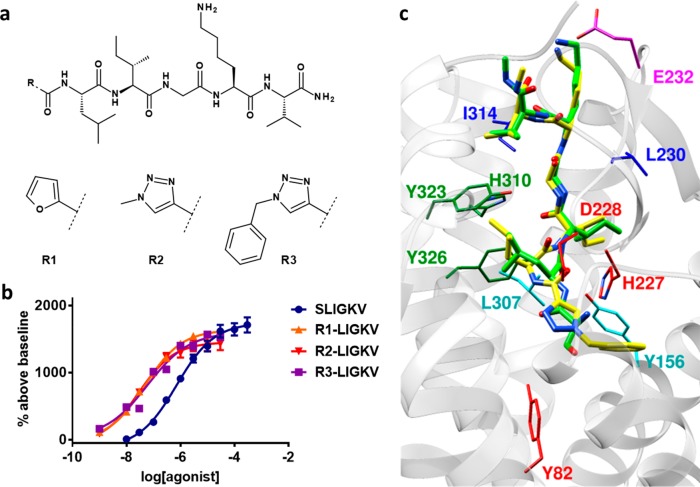
Analysis of the ligand-based shape alignment in relation to the PAR2 complex structure with AZ8838 suggested that there was an additional pocket in the receptor, occupied by the 4-fluoro-2-propylphenyl moiety of AZ8838, which was not utilized by SLIGKV or the agonist ligands. On the basis of this observation, a hybrid compound was designed to challenge the predicted binding mode by combining the residues 2–6 from the peptide sequence of SLIGKV with the benzyl-heterocyclic moiety of AZ8838 (Figure 7). A 2f-LIGKV compound (R1-LIGKV) analogous to 2f-LIGRLO-NH2 was obtained to confirm that the serine could be replaced by a heterocycle, whereas 1-methyl-1H[1,2,3]triazole-4-yl-LIGKV (R2-LIGKV) and 1-benzyl-1H[1,2,3]triazole-4-yl-LIGKV (R3-LIGKV) were synthesized to probe extensions into the novel pocket (Figure 8a). Replacing the N-terminus of SLIGKV with furan improved potency by 18-fold to pEC50 = 7.30 ± 0.06 (Figure 8b), in agreement with previous results for GB110 and 2f-LIGRLO-NH2.20,21,38 The use of a triazole moiety that extended further into the pocket with a methyl and benzyl group also resulted in highly potent agonists (pEC50 = 7.51 ± 0.06 and 7.30 ± 0.07 respectively, Figure 8b). Thus, our model facilitated the design of novel peptide agonists that were more potent than SLIGKV. This result also confirmed that the space identified in the model of SLIGKV bound to PAR2 could be exploited in agonist design and corroborates positioning of the N-terminus next to the occluded AZ8838 binding pocket. Considering that the rodent-derived peptide SLIGRL-NH2 is more potent than SLIGKV-NH2 at human PAR2,39 the potencies of the designed peptides could likely be further improved by replacing its lysine with an arginine.
Figure 8.
Design of novel agonists. (a) 2D structures of novel N-terminal modified peptide agonists. (b) Concentration-response curves of calcium signaling by novel agonists at PAR2. Data are presented as mean ± s.e.m. of n = 3 independent experiments. (c) Superimposed refined models of 1-benzyl-1H[1,2,3]triazole-4-yl-LIGKV (R3-LIGKV, yellow) and SLIGKV (green) from molecular docking calculations. Receptor residues have been color-coordinated based on their spatial arrangement.
Comparison of the binding modes obtained independently from the ligand-centric and molecular docking protocol showed that the same subpockets were explored by the N-terminus and hydrophobic side chains in positions 2 and 3 of SLIGKV (Figure S8). Docking of 1-benzyl-1H[1,2,3]triazole-4-yl-LIGKV to the receptor model obtained from the docking protocol resulted in poses consistent with the predictions made for the SLIGKV peptide (Figure 8c) as did those for GB110 and GB88 (Figure S9). In addition, docking of a series of peptides with modifications in the third position (SLMGKV, SL[Chg]GKV, and SLWGKV) resulted in poses consistent with that predicted for SLIGKV (Figures S10–S12). The cyclohexylglycine moiety of SL[Chg]GKV fitted neatly into the isoleucine pocket, whereas the larger tryptophan side chain of SLWGKV was more constrained due to clashes with L2094.60, Q214ECL2, and F2435.39, which was consistent with the lower activity of this substitution. The novel model of the agonist-bound state of PAR2 hence allowed docking of modified peptide agonists as well as nonpeptide agonists (GB110 and GB88), and the same binding mode was maintained throughout, providing a consistent hypothesis for agonist recognition. Unlike other GPCRs, the endogenous ligand of PAR2 is tethered, so there is no orthosteric ligand that can be used exogenously. This leads to difficulties in pharmacological characterization of the receptor in, for example, binding experiments or antagonism studies. The recognition that GB110 binds in a manner consistent with SLIGKV suggests that this small molecule tool compound can be used as a surrogate orthosteric ligand of PAR2 in future studies.
Novel Model of SLIGKV Bound to PAR2
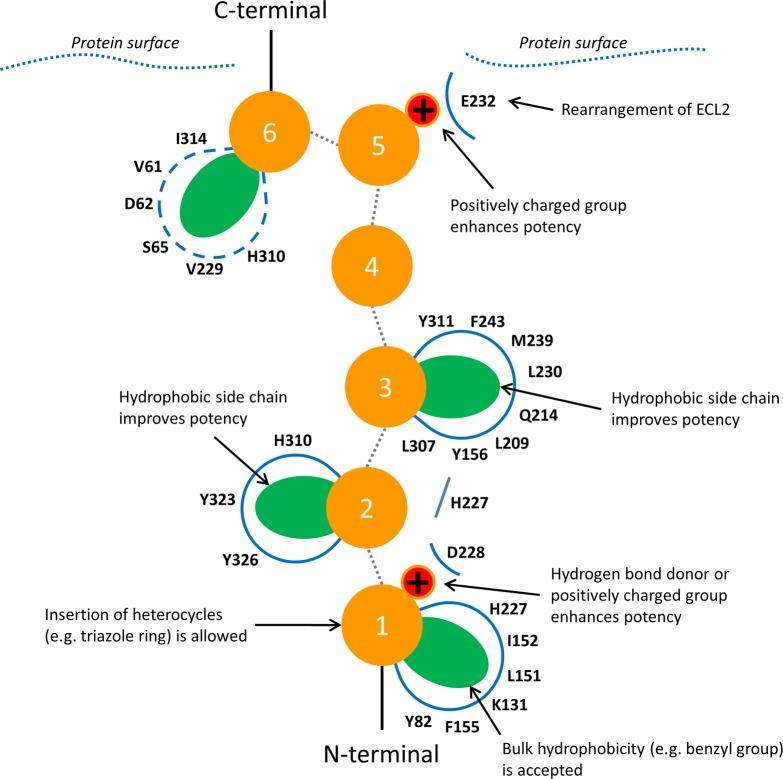
The publication of the first PAR2 crystal structure revealed the binding site of antagonist AZ8838 but was unable to identify the orthosteric binding site as it was not possible to crystallize complexes with agonist compounds. The novel model of agonist binding to PAR2 presented herein positions SLIGKV in the orthosteric site identified via mutagenesis (Figure 6). It provides structural insights into agonist-induced conformational changes, identifying that rearrangements of H227ECL2, E232ECL2, Y3237.32, Y3267.35, and H3106.58 in the antagonist-bound crystal structure are necessary to accommodate SLIGKV. The N-terminus of the peptide interacts with Y821.39 and D228ECL2, suggesting that the “gate-keeper” residue H227ECL2 must rearrange to allow the agonist to access the buried pocket accommodated by AZ8838. Cheng et al.16 proposed that AZ8838 occupies an allosteric site. Our study indicates that the compound binds in the orthosteric site, mimicking the interactions of the N-terminus of SLIGKV and thus acts as a competitive antagonist. The second residue of SLIGKV was predicted to be anchored in a pocket created by Y3237.32 and Y3267.35. Optimization of the interaction between the lysine to interact with E232ECL2 led to some repositioning of the third and fourth residues of SLIGKV. In our final model, a hydrophobic pocket for the third residue is delimited by M2395.35, Y1563.33, L2094.60, L230ECL2, F2435.39, L3076.55, and Y3116.59. The C-terminal valine of SLIGKV was predicted to bind in a pocket delimited by V61N-term, D62N-term, S65N-term, V229ECL2, Y3237.32, H3106.58, and I3146.62, but it should be noted that interactions in this region are expected to be dynamic. The key interactions made by the peptide in our final model are summarized in Figure 9.
Figure 9.
Predicted binding mode of peptide agonists in PAR2. Peptide residues are depicted as orange spheres and numbered according to their position. Hydrophobic (side chains shown as green ellipses) and charge-charge (red spheres) interactions were mapped through mutagenesis and modeling. Interactions of the residue at position 6 were not probed by mutagenesis and the region occupied by the side chain is expected to be dynamic.
In several previous studies, prediction of ligand-binding modes for PAR2 ligands was constrained to using homology models based on templates with low sequence identity, for example, bovine rhodopsin23,24,39 and human ORL-1 (nociceptin/orphanin FQ receptor)23,24,40 with TM sequence identities of 21 and 29%, respectively. Homology models based on these templates will have large errors in the ECL2 and relative orientation of TM helix 7, making it difficult to obtain an accurate structure of the binding site. The difficulties in obtaining a consistent binding mode for PAR2 agonists has been highlighted by a number of studies that used homology models based on templates with low sequence identity.24,39,40 Suen et al.24 recently identified residues important for 2f-LIGRL-NH2 binding based on mutagenesis (e.g., Y821.39, Y1563.33, D228ECL2, Y3116.59, Y3267.35, and L3307.39) and primarily docked this compound to a rhodopsin-based homology model to identify potential binding modes. Although experimentally the study found a similar set of residues to be important for activation of PAR2, the binding mode predicted for 2f-LIGRL-NH2 is fundamentally different from that proposed in this study for SLIGKV and similar synthetic agonists. As demonstrated by the ligand-based alignments made in this study, the similarities between SLIGKV and 2f-LIGRL-NH2 suggest that these agonists should have a conserved binding mode. The present model predicted that the N-terminus is positioned close to both Y821.39 and D228ECL2 whereas these residues appear to be located in distinct pockets in the homology model based on rhodopsin.24 Due to differences in ECL2 of the rhodopsin-based model, D228ECL2 is facing into the extracellular space and was previously predicted to interact with the arginine residue of 2f-LIGRL-NH2 (equivalent to the lysine of SLIGKV). In addition, the C-terminal leucine of 2f-LIGRL-NH2 (corresponding to the valine in SLIGKV) was predicted to interact with Y821.39, whereas we position the N-terminal moiety in this region. The former option was obtained as one of our docking solutions in the PAR2 structure, but it was discarded because of the constraint that the C-terminus of the peptide must extend to the extracellular space to be consistent with the endogenous tethered ligand. The PAR1 crystal structure provided a better template for PAR2 homology modeling,24 based on high TM sequence identity (44%), but the binding site conformation may not be relevant for PAR2 as it was determined in complex with a PAR1 selective antagonist (vorapaxar). In addition, a homology model based on PAR1 differs from the PAR2 crystal structure in ECL2 and TM helices 5 and 6. Therefore, even though the PAR2 crystal structure was also determined in an inactive conformation,16 it represented an improved starting point for modeling. The structure brought the occluded binding pocket occupied by AZ8838 to our attention and demonstrated that D228ECL2 could form key interactions in this region. However, it should be emphasized that the crystal structure alone provided few clues on the interactions with SLIGKV. The constraints derived from the mutations and modified peptides were crucial in the modeling process as docking poses were selected based on these experiments.
In light of the new PAR2 crystal structures, an extensive array of mutagenesis data generated for this GPCR has been placed into structural context for the first time. In support of the previous studies on modification of peptide agonists, we report the need for a charged or heterocyclic moiety at the amino terminus,17,41 the importance of the leucine at position 2,27 and improved potencies for larger hydrophobic side chains (e.g., Cha and Chg) at positions 2 and 3.26 Our experimental data also align well with other receptor mutagenesis studies which are summarized in Figure S13 and Table S3. In agreement with Suen et al.,24 mutations of 5 receptor residues (Y821.39, Y1563.33, D228ECL2, E232ECL2, and Y3267.35) were detrimental, suggesting these are key for activation of calcium signaling. ECL2 has been reported to be important for peptide recognition at PAR2 in several studies,28,29,42 and we identified potential interaction partners in SLIGKV. The predicted structure of the PAR2–SLIGKV complex represents an agonist-bound state, in which the receptor backbone is in an overall inactive conformation. This static model cannot explain how the changes induced by binding of the peptide propagate toward the intracellular side but could provide a starting point for understanding PAR2-mediated activation of calcium and other signaling pathways at the molecular level. Future studies should consider effects of mutants located deeper in the transmembrane region and the influence of the unique activation mechanism of PAR2 on signaling. Notably, all receptor mutations had much smaller effects when activated by the protease compared to the synthetic peptide. This phenomenon has also been detected at PAR131 and could be because the tethered ligand has a larger interaction site across the surface of the receptor, which could affect receptor signaling. In fact, several studies have highlighted that there may be differences in activation of PAR2-mediated signaling via calcium and MAP kinase pathways by the tethered ligand compared to peptides.25,27,43 To fully address this conundrum, future work should consider activation of multiple pathways as well as agonists with biased signaling properties.7,44,45
Conclusions
Drug discovery for PARs has been hampered by the challenges involved in development of ligands. Access to structural information and understanding of the interactions responsible for receptor activation provides opportunities for rational drug design. Our model of agonist-bound PAR2 shows that SLIGKV binds in an extended conformation, occupying two pockets identified in the inactive crystal structure. The same binding mode was maintained in models of synthetic agonists, which supports that interactions in both sites are necessary for receptor activation and guided design of new PAR2 agonists. This also revealed the structural basis of antagonism by AZ8838, highlighting that the antagonist blocks access to the region of the orthosteric site that recognizes the N-terminus of SLIGKV. Antagonists may be obtained by targeting the AZ8838 pocket, but as optimization of potency may be challenging due to its limited size, the novel orthosteric site identified in this work provides an attractive alternative. The model could be useful in future virtual screening campaigns for novel PAR2 antagonists and agonists and in design of experiments focused on understanding the structural basis of receptor activation. The combined experimental and computational approach to characterize agonist binding modes can be extended to study the many other GPCRs that recognize peptides or proteins.
Methods
Materials
PAR2 was expressed in the recombinant 1321N1 cell line (ATCC). Plasmids containing PAR2 with/without single point mutations were purchased from ThermoFisher. SLIGKV and mutant peptide agonists (2D structures can be found in Figure S1) were purchased from ThermoFisher except SL[OtBu]GKV and S[Phe(3,4-diChloro)]IGKV, which were purchased from Chinese Peptide Company, China. All peptides were amidated at the C-terminus. All peptides were >95% pure. Small molecule GB110 was synthesized as in Cheng et al.16
Design of Point Mutations
In addition to previously available mutants of PAR216 covering the occluded antagonist pocket (I327L, H135Y, H227A, H227Q, D228A, and D228N), reports of important amino acids from the literature30,42 as well as residues lining the channel from the extracellular region toward the occluded binding site were selected (L230ECL2, E232ECL2.28, Q233ECL2, L3076.55, H3106.58, L3146.62, and L3307.39). For Y1563.33, Y3116.59, Y3237.32, and Y3267.35, both the alanine and the phenylalanine mutants were investigated to explore the impact of a large modification compared to removing the hydrogen bond capacity. The Y82F mutant was constructed to probe the importance of a hydrogen bond observed in the complex structure of PAR2–AZ8838. To further evaluate the effects of these receptor mutations, complementary point mutations were made in the SLIGKV peptide sequence based on initial hypotheses of how the peptide would interact with the receptor as well as previous studies.26,41
Transient Transfection of PAR2 Mutant Receptors
1321N1 cells were cultured in Dulbecco’s modified Eagle’s medium with glutamax-1 (DMEM) containing 10% fetal bovine serum (FBS). Cells were seeded at around 40 000 cells/cm3 the day before transfection. Plasmid DNA was heat inactivated at 65 °C for 20 min prior to electroporation. Transient transfections of PAR2 were carried out using the MaxCyte STX scalable transfection system, following the standard protocol with the subsequent modifications. Cells were detached, washed, and resuspended at 1 × 108 cells/mL in MaxCyte electroporation buffer. Plasmid DNA (50 μg/mL of cells) was prepared. Cells (400 μL) were added to the DNA, mixed thoroughly and transferred to the OC-400 cuvette. Electroporation was performed using the protocol appropriate for 1321N1 cells, and the cells were then allowed to recover at 37 °C for 15 min. The transfected cells were then cryopreserved in assay ready vials.
Calcium Mobilization Assay
PAR2-induced calcium release was monitored using the Screen Quest Fluo-8 No Wash Calcium Assay Kit (AAT Bioquest), following the manufacturer’s protocol with the subsequent modifications. Parental and PAR2 transiently transfected 1321N1 cells were seeded in DMEM containing 10% FBS at 4000 cells/well/20 μL in 384-well plates (Corning #3770) and incubated overnight at 37 °C, 5% CO2. Agonists were prepared in HHBS buffer (1× HBSS, 20 mM HEPES pH 7.4) containing 0.1% BSA, at 5 times the final assay concentration (stated in parentheses): Trypsin (0.3 μM to 30 pM); SLIGKV and modified peptides (1 mM to 30 nM) except SLIGEV, SLIGAV (0.3 mM to 30 nM). Cells were loaded with Fluo-8 dye loading solution and incubated at 37 °C, 5% CO2 for 30 min, and then at room temperature for 30 min. The calcium assay was then run in a FLIPR tetra high-throughput cellular screening system (Molecular Devices) by monitoring the fluorescence intensity at Ex/Em = 490/525 nm. In the case of trypsin, after 30 min of incubation of the cells with the Fluo-8 dye, DMSO (0.01%) or vorapaxar (1 μM) was added for 30 min of incubation at room temperature prior addition of agonist. This was to ensure that the trypsin response monitored was PAR2 specific. Addition of DMSO/vorapaxar prior to a single concentration of SLIGKV (10 μM) showed that vorapaxar did not interact with the mutant PAR2 receptors. Individual concentration-response curves were plotted in RLU using GraphPad Prism 746 and the “log[agonist] vs response-variable slope (four parameter)” curve fit to obtain values of potency, EC50 (concentration producing half maximal response) and efficacy, Emax (maximum response). Responses were then normalized as a percentage above the baseline measurement on the corresponding plate and combined as appropriate. EC50 data were converted to pEC50 (−log10 EC50) and collated to give a mean value. Results are expressed as mean ± standard error of the mean (s.e.m.) of n independent experiments as stated in the table/figure legends. For each receptor, pEC50 for the mutated peptides were compared to that of SLIGKV, and for each peptide, pEC50 for the mutated receptors were compared to WT receptor. To better illustrate differences between peptides and/or receptors, the fold change was calculated based on the geometric means (EC50 mutant/EC50 WT). If no response was obtained functionally (i.e., pEC50 is less than the highest concentration tested), then the minimum possible value was used in place of the geometric mean and the fold change is expressed as greater than.
Molecular Docking Calculations
The crystal structure of human PAR2 in complex with AZ8838 (PDB ID: 5NDD)16 was prepared for docking simulations: (1) Solvent and buffer molecules as well as T4 lysozyme and cytochrome b562RIL insertions were removed. (2) N- and C-termini were capped with acetyl (ACE) and N-methyl amide (NME) groups respectively. (3) Hydrogens were added to the complex. (4) AZ8838 was finally removed. Structures of peptide agonists (SLIGKV, 1-benzyl-1H[1,2,3]triazole-4-yl-LIGKV, SLMGKV, SLWGKV, and SL[Chg]GKV) were built, capped with NME, and protonated at physiological pH using PyMol.47 Structures of GB88 and GB110 were built in Marvin,48 and low-energy conformers were obtained using the cxcalc tool. A total of 500 conformers were generated using the hyperfine option and optimization limit set to 2.
AutoDock33 (release 4.2.6) was compiled to handle >32 rotatable bonds. Receptor flexibility was accounted for by considering multiple side chain rotamers for H3106.58, Y3116.59, Y3237.32, Y3267.35, H227ECL2, and Q233ECL2 during the docking calculations. Docking input files were prepared through the AutoDockTools (ADT) package33 using a grid of 59 × 67 × 51 points in the xyz dimension with a spacing of 0.375 Å. AutoGrid4 was used to generate grid maps. The Lamarckian genetic algorithm (LGA) was employed: The global optimization started with a population of 300 individuals; a maximum of 10 million energy evaluations, 27 000 generations, and 100 runs were set. Then, 100 independent screens were performed to collect a total of 10 000 poses. The results from AutoDock were collected to perform a hierarchical cluster analysis for the 10 000 poses. Clustering was performed by means of the Clusterizer 1.1 software49 implemented with the compute_rms_between_conformation.py function from the AutoDockTools (ADT) package.33
Complex Refinement
MODELLER 9.1435 was used to refine the model obtained from flexible receptor docking. Loop modeling refinement was applied to optimize: (1) SLIGKV; (2) the series of side chains considered as flexible during docking (H3106.58, Y3116.59, Y3237.32, Y3267.35, and H227ECL2); (3) the protein sequence between L230ECL2–L234ECL2 to facilitate salt bridge interaction between the lysine and E232ECL2 in ECL2. A total of 250 models were generated, sorted by DOPE score, and visually inspected.
Docking and Refinement of PAR2–1-Benzyl-1H[1,2,3]triazole-4-yl-LIGKV, −SLMGKV, −SLWGKV, −SL[Chg]GKV, −GB110, and −GB88
1-benzyl-1H[1,2,3]triazole-4-yl-LIGKV, SLMGKV, SLWGKV, SL[Chg]GKV, GB110, and GB88 were docked into the protein structure of the refined PAR2–SLIGKV model. In contrast to the docking of SLIGKV, the protein was considered rigid during the calculations, and the total number of collected poses was reduced to 100. Cluster analysis was performed by considering a standard RMSD threshold of 2 Å between the poses obtained using docking. Except for GB110 and GB88, MODELLER 9.1435 was used to refine the selected model by optimizing only the residues of the peptide. A total of 25 models were generated, sorted by DOPE score and visually inspected.
Ligand-Based Modeling
Conformational searches of GB88 and truncated tetrapeptide versions of SLIGKV (SLIG) as well as 2f-LIGRLO-NH2 (2f-LIG) were performed using MacroModel-v11.350 using the OPLS3 force field with implicit (water) solvent, as implemented in Maestro.51 Ligand-based shape overlay was performed using ROCS 3.2.0.436,52 with AZ3833 and added carbon atoms in the hypothesized pocket for residues 2 and 3 as a query. A refined protein model was constructed from the PAR2 sequence using Prime53 and 5NDD16 as a template with the GB88 model added to the protein template to allow for side chain rearrangement.
Images
Molecular graphics were generated using the UCSF Chimera package,54 and chemical structures were drawn in BIOVIA Draw 16.1.55
Chemical Synthesis of N-Terminal Modifications of SLIGKV
The LIGKV sequence was set up and run on a Biotage Alstra SPPS synthesizer using the following method: The resin (0.65 g, 0.33 mmol, 0.50 mmol/g) was swelled in DMF at 55 °C. Fmoc deprotection was carried out in 20% Pip in DMF 2 × (3 + 10 min) followed by DMF wash (5×). Amide coupling (0.2 M amino acid in DMF, 4 equiv) using DIC (2 M in DMF)/Oxyma (0.5 M in DMF) was run at 75 °C for 7 min followed by DMF wash (5×). The deprotection and coupling was repeated for every amino acid. The final Fmoc deprotection was carried out in 20% Pip in DMF 2 × (3 + 10 min), and the resin was washed with DMF (5×), MeOH (5×), and DCM (5×). Test cleavage for 40 min in TFA/Water/TiPS (95:2.5:2.5) showed product by LCMS. The resin was split into four solid phase synthesis reactor vials.
N-Terminal carboxylic acids (0.33 mmol) and HATU (125 mg, 0.33 mmol) were suspended in DMF (3 mL), and DIPEA (0.1 mL, 0.57 mmol) was added. When a clear solution was obtained it was added to the resin (0.082 mmol), and the reaction mixtures was shaken for 1.5 h. The solutions were filtered off, and the resins were washed with DMF (5 × 5 mL) followed by MeOH (3 × 5 mL). Cleavage from the resin was carried out using a pre-cooled solution of TFA/Water/TiPS (95:2.5:2.5, ∼5 mL) and agitated for 1 h 20 min. The cleavage mixtures were concentrated, precipitated from DEE, and centrifuged, and the ether layers were decanted off. The precipitates were redissolved in water/acetonitrile/acetic acid (68:30:2) and lyophilized to obtain the crude peptides. The crude peptides were purified by HPLC (on a C18 column, 150 × 19 mm, 5 μm particles, using a 20 min gradient of acetonitrile in 0.15% TFA buffer), and the pure fraction was lyophilized with 2% AcOH twice to obtain the modified peptides as TFA salts.
N-((3S,6S,12S,15S)-6-(4-Aminobutyl)-12-((S)-sec-butyl)-3-carbamoyl-2,17-dimethyl-5,8,11,14-tetraoxo-4,7,10,13-tetraazaoctadecan-15-yl)-1-methyl-1H-1,2,3-triazole-4-carboxamide
White solid. Yield 12.4 mg (20%). 1H NMR (500 MHz, DMSO-d6) δ: 8.53 (s, 1H), 8.44 (d, J = 8.9 Hz, 1H), 8.27 (t, J = 5.6 Hz, 1H), 7.93 (dd, J = 11.0, 8.5 Hz, 2H), 7.72 (d, J = 8.9 Hz, 1H), 7.68 (s, 3H), 7.39 (s, 1H), 7.03 (s, 1H), 4.58 (td, J = 10.0, 9.8, 4.7 Hz, 1H), 4.33 (q, J = 8.1 Hz, 1H), 4.20 (t, J = 7.9 Hz, 1H), 4.09 (s, 3H), 4.04–4.08 (m, 1H), 3.79 (dd, J = 16.5, 5.7 Hz, 1H), 3.65 (dd, J = 16.6, 5.6 Hz, 1H), 2.69–2.79 (m, 2H), 1.91–1.98 (m, 1H), 1.55–1.75 (m, 4H), 1.40–1.55 (m, 5H), 1.22–1.35 (m, 2H), 1.03–1.15 (m, 1H), 0.87 (dd, J = 9.2, 6.5 Hz, 6H), 0.78–0.85 (m, 12H). HRMS (ESI-QTOF): m/z [M + H]+ Calcd for C29H53N10O6 637.4144. Found 637.4180.
N-((3S,6S,12S,15S)-6-(4-Aminobutyl)-12-((S)-sec-butyl)-3-carbamoyl-2,17-dimethyl-5,8,11,14-tetraoxo-4,7,10,13-tetraazaoctadecan-15-yl)furan-2-carboxamide
White solid. Yield 15.3 mg (25%). 1H NMR (500 MHz, DMSO-d6) δ: 8.34 (d, J = 8.5 Hz, 1H), 8.25 (t, J = 5.7 Hz, 1H), 7.94 (d, J = 8.0 Hz, 1H), 7.86 (d, J = 0.9 Hz, 1H), 7.83 (d, J = 8.6 Hz, 1H), 7.73 (d, J = 8.9 Hz, 1H), 7.70 (s, 3H), 7.39 (s, 1H), 7.18 (d, J = 2.9 Hz, 1H), 7.03 (s, 1H), 6.63 (dd, J = 3.5, 1.7 Hz, 1H), 4.47–4.56 (m, 1H), 4.29–4.37 (m, 1H), 4.18 (t, J = 8.0 Hz, 1H), 4.08 (dd, J = 8.9, 6.6 Hz, 1H), 3.79 (dd, J = 16.6, 5.8 Hz, 1H), 3.65 (dd, J = 16.5, 5.6 Hz, 1H), 2.69–2.79 (m, 2H), 1.91–1.99 (m, 1H), 1.56–1.75 (m, 4H), 1.41–1.56 (m, 5H), 1.25–1.35 (m, 2H), 1.02–1.13 (m, 1H), 0.88 (d, J = 6.5 Hz, 3H), 0.78–0.87 (m, 15H). HRMS (ESI-QTOF): m/z [M + H]+ Calcd for C30H52N7O7 622.3923. Found 622.3937.
N-((3S,6S,12S,15S)-6-(4-Aminobutyl)-12-((S)-sec-butyl)-3-carbamoyl-2,17-dimethyl-5,8,11,14-tetraoxo-4,7,10,13-tetraazaoctadecan-15-yl)-1-benzyl-1H-1,2,3-triazole-4-carboxamide
White solid. Yield 21.2 mg (30%). 1H NMR (500 MHz, DMSO-d6) δ: 8.70 (s, 1H), 8.46 (d, 1H), 8.27 (t, 1H), 7.89–8.00 (m, 2H), 7.72 (d, 1H), 7.66 (s, 3H), 7.32–7.44 (m, 6H), 7.04 (s, 1H), 5.66 (s, 2H), 4.53–4.64 (m, 1H), 4.28–4.38 (m, 1H), 4.19 (t, 1H), 4.09 (t, 1H), 3.73–3.84 (m, 1H), 3.61–3.70 (m, 1H), 2.69–2.80 (m, 2H), 1.92–1.98 (m, 1H), 1.61–1.75 (m, 3H), 1.42–1.61 (m, 6H), 1.25–1.35 (m, 2H), 1.03–1.13 (m, 1H), 0.76–0.91 (m, 18H). HRMS (ESI-QTOF): m/z [M + H]+ Calcd for C35H57N10O6 713.4457. Found 713.4489.
(S)-6-Amino-N-((S)-1-amino-3-methyl-1-oxobutan-2-yl)-2-(2-((2S,3S)-2-((S)-2-((S)-3-hydroxy-2-methylpropanamido)-4-methylpentanamido)-3-methylpentanamido)acetamido)hexanamide
White solid. Yield 9.7 mg (16%). 1H NMR (500 MHz, DMSO-d6) δ 8.20 (t, J = 5.7 Hz, 1H), 8.12 (d, J = 8.1 Hz, 1H), 7.92 (d, J = 8.1 Hz, 1H), 7.71 (d, J = 8.8 Hz, 4H), 7.62 (d, J = 8.5 Hz, 1H), 7.39 (s, 1H), 7.04 (s, 1H), 4.88 (s, 1H), 4.30–4.37 (m, 1H), 4.27 (q, J = 7.7 Hz, 1H), 4.13 (t, J = 7.7 Hz, 1H), 4.09 (dd, J = 6.8, 8.7 Hz, 1H), 3.65–3.80 (m, 2H), 3.33–3.50 (m, 2H, one proton under water signal), 2.69–2.81 (m, 2H), 2.48–2.53 (m, 1H, one proton under DMSO signal), 1.93–2.00 (m, 1H), 1.69–1.76 (m, 1H), 1.64–1.69 (m, 1H), 1.57–1.64 (m, 1H), 1.46–1.56 (m, 5H), 1.38–1.46 (m, 1H), 1.25–1.35 (m, 2H), 1.02–1.12 (m, 1H), 0.94 (d, J = 6.8 Hz, 3H), 0.78–0.90 (m, 18H). (two protons under solvent signals assigned from COSEY). HRMS (ESI-QTOF): m/z [M + H]+ Calcd for C29H56N7O7 614.4236. Found 614.4256.
Acknowledgments
A.J.K is a fellow of the AstraZeneca postdoc programme. We acknowledge support at AstraZeneca from N. O. Hermansson for contributions in receptor transfection, E. Gordon for design of plasmid constructs, the Physical and Analytical Chemistry team in RIA IMED Biotech Unit for HRMS analysis, and N. Dekker for helpful discussions throughout. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement: 715052). The work was also supported by grants from the Science for Life Laboratory and the Swedish Research Council (2017-4676) to J.C. The computations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at C3SE and NSC.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsptsci.8b00019.
Author Contributions
⊥ A.J.K. and F.B. contributed equally to this work.
The authors declare no competing financial interest.
This article is made available for a limited time sponsored by ACS under the ACS Free to Read License, which permits copying and redistribution of the article for non-commercial scholarly purposes.
Supplementary Material
References
- Mackie E. J.; Pagel C. N.; Smith R.; de Niese M. R.; Song S. J.; Pike R. N. (2002) Protease-activated receptors: a means of converting extracellular proteolysis into intracellular signals. IUBMB Life 53, 277–281. 10.1080/15216540213469. [DOI] [PubMed] [Google Scholar]
- Nystedt S.; Emilsson K.; Wahlestedt C.; Sundelin J. (1994) Molecular cloning of a potential proteinase activated receptor. Proc. Natl. Acad. Sci. U. S. A. 91, 9208–9212. 10.1073/pnas.91.20.9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T.; Hirano K.; Hirano M.; Nishimura J.; Kuga H.; Nakamura K.; Takahashi S.; Kanaide H. (2004) Inactivation of protease-activated receptor-1 by proteolytic removal of the ligand region in vascular endothelial cells. Biochem. Pharmacol. 68, 23–32. 10.1016/j.bcp.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Nakayama T.; Hirano K.; Shintani Y.; Nishimura J.; Nakatsuka A.; Kuga H.; Takahashi S.; Kanaide H. (2003) Unproductive cleavage and the inactivation of protease-activated receptor-1 by trypsin in vascular endothelial cells. Br. J. Pharmacol. 138, 121–130. 10.1038/sj.bjp.0705008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole R. M.; Elkinson S. (2014) Vorapaxar: first global approval. Drugs 74, 1153–1163. 10.1007/s40265-014-0252-2. [DOI] [PubMed] [Google Scholar]
- Geppetti P.; Veldhuis N. A.; Lieu T.; Bunnett N. W. (2015) G Protein-Coupled Receptors: Dynamic Machines for Signaling Pain and Itch. Neuron 88, 635–649. 10.1016/j.neuron.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Zhao P.; Lieu T.; Barlow N.; Sostegni S.; Haerteis S.; Korbmacher C.; Liedtke W.; Jimenez-Vargas N. N.; Vanner S. J.; Bunnett N. W. (2015) Neutrophil Elastase Activates Protease-activated Receptor-2 (PAR2) and Transient Receptor Potential Vanilloid 4 (TRPV4) to Cause Inflammation and Pain. J. Biol. Chem. 290, 13875–13887. 10.1074/jbc.M115.642736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.; Yau M. K.; Lim J.; Wu K. C.; Xu W.; Suen J. Y.; Fairlie D. P. (2018) A Potent Antagonist of Protease-Activated Receptor 2 That Inhibits Multiple Signaling Functions in Human Cancer Cells. J. Pharmacol. Exp. Ther. 364, 246–257. 10.1124/jpet.117.245027. [DOI] [PubMed] [Google Scholar]
- Ungefroren H.; Witte D.; Rauch B. H.; Settmacher U.; Lehnert H.; Gieseler F.; Kaufmann R. (2017) Proteinase-Activated Receptor 2 May Drive Cancer Progression by Facilitating TGF-beta Signaling. Int. J. Mol. Sci. 18, 2494–2507. 10.3390/ijms18112494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg M. D.; Mihara K.; Polley D.; Suen J. Y.; Han A.; Fairlie D. P.; Ramachandran R. (2014) Biased signalling and proteinase-activated receptors (PARs): targeting inflammatory disease. Br. J. Pharmacol. 171, 1180–1194. 10.1111/bph.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmeier A. S.; Ruf W. (2012) Protease-activated receptor 2 signaling in inflammation. Semin. Immunopathol. 34, 133–149. 10.1007/s00281-011-0289-1. [DOI] [PubMed] [Google Scholar]
- Kagota S.; Maruyama K.; McGuire J. J. (2016) Characterization and Functions of Protease-Activated Receptor 2 in Obesity, Diabetes, and Metabolic Syndrome: A Systematic Review. BioMed Res. Int. 2016, 1. 10.1155/2016/3130496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci M.; Roviezzo F.; Cirino G. (2005) Protease-activated receptor-2 (PAR2) in cardiovascular system. Vasc. Pharmacol. 43, 247–253. 10.1016/j.vph.2005.07.009. [DOI] [PubMed] [Google Scholar]
- McGuire J. J. (2004) Proteinase-Activated Receptor 2 (PAR2): A Challenging New Target for Treatment of Vascular Diseases. Curr. Pharm. Des. 10, 2769–2778. 10.2174/1381612043383656. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Srinivasan Y.; Arlow D. H.; Fung J. J.; Palmer D.; Zheng Y.; Green H. F.; Pandey A.; Dror R. O.; Shaw D. E.; Weis W. I.; Coughlin S. R.; Kobilka B. K. (2012) High-resolution crystal structure of human protease-activated receptor 1. Nature 492, 387–392. 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R. K. Y.; Fiez-Vandal C.; Schlenker O.; Edman K.; Aggeler B.; Brown D. G.; Brown G. A.; Cooke R. M.; Dumelin C. E.; Dore A. S.; Geschwindner S.; Grebner C.; Hermansson N. O.; Jazayeri A.; Johansson P.; Leong L.; Prihandoko R.; Rappas M.; Soutter H.; Snijder A.; Sundstrom L.; Tehan B.; Thornton P.; Troast D.; Wiggin G.; Zhukov A.; Marshall F. H.; Dekker N. (2017) Structural insight into allosteric modulation of protease-activated receptor 2. Nature 545, 112–115. 10.1038/nature22309. [DOI] [PubMed] [Google Scholar]
- Hollenberg M. D.; Saifeddine M.; Al-Ani B.; Kawabata A. (1997) Proteinase-activated receptors: structural requirements for activity, receptor cross-reactivity, and receptor selectivity of receptor-activating peptides. Can. J. Physiol. Pharmacol. 75, 832–841. 10.1139/y97-110. [DOI] [PubMed] [Google Scholar]
- Kanke T.; Ishiwata H.; Kabeya M.; Saka M.; Doi T.; Hattori Y.; Kawabata A.; Plevin R. (2005) Binding of a highly potent protease-activated receptor-2 (PAR2) activating peptide, [3H]2-furoyl-LIGRL-NH2, to human PAR2. Br. J. Pharmacol. 145, 255–263. 10.1038/sj.bjp.0706189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg M. D.; Renaux B.; Hyun E.; Houle S.; Vergnolle N.; Saifeddine M.; Ramachandran R. (2008) Derivatized 2-furoyl-LIGRLO-amide, a versatile and selective probe for proteinase-activated receptor 2: binding and visualization. J. Pharmacol. Exp. Ther. 326, 453–462. 10.1124/jpet.108.136432. [DOI] [PubMed] [Google Scholar]
- Barry G. D.; Suen J. Y.; Le G. T.; Cotterell A.; Reid R. C.; Fairlie D. P. (2010) Novel agonists and antagonists for human protease activated receptor 2. J. Med. Chem. 53, 7428–7440. 10.1021/jm100984y. [DOI] [PubMed] [Google Scholar]
- Suen J. Y.; Barry G. D.; Lohman R. J.; Halili M. A.; Cotterell A. J.; Le G. T.; Fairlie D. P. (2012) Modulating human proteinase activated receptor 2 with a novel antagonist (GB88) and agonist (GB110). Br. J. Pharmacol. 165, 1413–1423. 10.1111/j.1476-5381.2011.01610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufareva I.; Rueda M.; Katritch V.; Stevens R. C.; Abagyan R. (2011) Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure 19, 1108–1126. 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S. R.; Xu W.; Wirija A.; Lim J.; Yau M. K.; Stoermer M. J.; Lucke A. J.; Fairlie D. P. (2015) Three Homology Models of PAR2 Derived from Different Templates: Application to Antagonist Discovery. J. Chem. Inf. Model. 55, 1181–1191. 10.1021/acs.jcim.5b00087. [DOI] [PubMed] [Google Scholar]
- Suen J. Y.; Adams M. N.; Lim J.; Madala P. K.; Xu W.; Cotterell A. J.; He Y.; Yau M. K.; Hooper J. D.; Fairlie D. P. (2017) Mapping transmembrane residues of proteinase activated receptor 2 (PAR2) that influence ligand-modulated calcium signaling. Pharmacol. Res. 117, 328–342. 10.1016/j.phrs.2016.12.020. [DOI] [PubMed] [Google Scholar]
- Ramachandran R.; Mihara K.; Mathur M.; Rochdi M. D.; Bouvier M.; Defea K.; Hollenberg M. D. (2009) Agonist-biased signaling via proteinase activated receptor-2: differential activation of calcium and mitogen-activated protein kinase pathways. Mol. Pharmacol. 76, 791–801. 10.1124/mol.109.055509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryanoff B. E.; Santulli R. J.; McComsey D. F.; Hoekstra W. J.; Hoey K.; Smith C. E.; Addo M.; Darrow A. L.; Andrade-Gordon P. (2001) Protease-activated receptor-2 (PAR-2): structure-function study of receptor activation by diverse peptides related to tethered-ligand epitopes. Arch. Biochem. Biophys. 386, 195–204. 10.1006/abbi.2000.2207. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Yau M. K.; Kok W. M.; Lim J.; Wu K. C.; Liu L.; Hill T. A.; Suen J. Y.; Fairlie D. P. (2017) Biased Signaling by Agonists of Protease Activated Receptor 2. ACS Chem. Biol. 12, 1217–1226. 10.1021/acschembio.6b01088. [DOI] [PubMed] [Google Scholar]
- Al-Ani B.; Wijesuriya S. J.; Hollenberg M. D. (2002) Proteinase-activated receptor 2: differential activation of the receptor by tethered ligand and soluble peptide analogs. J. Pharmacol. Exp. Ther. 302, 1046–1054. 10.1124/jpet.302.3.1046. [DOI] [PubMed] [Google Scholar]
- Lerner D. J.; Chen M.; Tram T.; Coughlin S. R. (1996) Agonist recognition by proteinase-activated receptor 2 and thrombin receptor—importance of extracellular loop interactions for receptor function. J. Biol. Chem. 271, 13943–13947. 10.1074/jbc.271.24.13943. [DOI] [PubMed] [Google Scholar]
- Al-Ani B.; Saifeddine M.; Kawabata A.; Hollenberg M. D. (1999) Proteinase activated receptor 2: role of extracellular loop 2 for ligand-mediated activation. Br. J. Pharmacol. 128, 1105–1113. 10.1038/sj.bjp.0702834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhart B. D.; Ruslim-Litrus L.; Lu C. C.; Alves V. L.; Teng W.; Scarborough R. M.; Reynolds E. E.; Oksenberg D. (2000) Extracellular mutations of protease-activated receptor-1 result in differential activation by thrombin and thrombin receptor agonist peptide. Mol. Pharmacol. 58, 1178–1187. 10.1124/mol.58.6.1178. [DOI] [PubMed] [Google Scholar]
- Isberg V.; de Graaf C.; Bortolato A.; Cherezov V.; Katritch V.; Marshall F. H.; Mordalski S.; Pin J. P.; Stevens R. C.; Vriend G.; Gloriam D. E. (2015) Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 36, 22–31. 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanevicz T. I.M.; Ishii M.; Wang L.; Chen M.; Chen J.; Turck C. W.; Cohen F. E.; Coughlin S. R. (1995) Mechanisms of thrombin receptor agonist specificity. Chimeric receptors and complementary mutations identify an agonist recognition site. J. Biol. Chem. 270, 21619–21625. 10.1074/jbc.270.37.21619. [DOI] [PubMed] [Google Scholar]
- Sali A.; Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815. 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Hawkins P. C. D.; Skillman A. G.; Nicholls A. (2007) Comparison of Shape-Matching and Docking as Virtual Screening Tools. J. Med. Chem. 50, 74–82. 10.1021/jm0603365. [DOI] [PubMed] [Google Scholar]
- Jacobson M. P.; Kaminski G. A.; Friesner R. A.; Rapp C. S. (2002) Force Field Validation Using Protein Side Chain Prediction. J. Phys. Chem. B 106, 11673–11680. 10.1021/jp021564n. [DOI] [Google Scholar]
- McGuire J. J.; Saifeddine M.; Triggle C. R.; Sun K.; Hollenberg M. D. (2004) 2-furoyl-LIGRLO-amide: a potent and selective proteinase-activated receptor 2 agonist. J. Pharmacol. Exp. Ther. 309, 1124–1131. 10.1124/jpet.103.064584. [DOI] [PubMed] [Google Scholar]
- Yau M. K.; Liu L.; Fairlie D. P. (2013) Toward drugs for protease-activated receptor 2 (PAR2). J. Med. Chem. 56, 7477–7497. 10.1021/jm400638v. [DOI] [PubMed] [Google Scholar]
- Yau M. K.; Suen J. Y.; Xu W.; Lim J.; Liu L.; Adams M. N.; He Y.; Hooper J. D.; Reid R. C.; Fairlie D. P. (2016) Potent Small Agonists of Protease Activated Receptor 2. ACS Med. Chem. Lett. 7, 105–110. 10.1021/acsmedchemlett.5b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G. D.; Suen J. Y.; Low H. B.; Pfeiffer B.; Flanagan B.; Halili M.; Le G. T.; Fairlie D. P. (2007) A refined agonist pharmacophore for protease activated receptor 2. Bioorg. Med. Chem. Lett. 17, 5552–5557. 10.1016/j.bmcl.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Ma J. N.; Burstein E. S. (2013) The protease activated receptor 2 (PAR2) polymorphic variant F240S constitutively activates PAR2 receptors and potentiates responses to small-molecule PAR2 agonists. J. Pharmacol. Exp. Ther. 347, 697–704. 10.1124/jpet.113.208744. [DOI] [PubMed] [Google Scholar]
- Al-Ani B.; Hansen K. K.; Hollenberg M. D. (2004) Proteinase-Activated Receptor-2: Key Role of Amino-Terminal Dipeptide Residues of the Tethered Ligand for Receptor Activation. Mol. Pharmacol. 65, 149–156. 10.1124/mol.65.1.149. [DOI] [PubMed] [Google Scholar]
- Elmariah S. B.; Reddy V. B.; Lerner E. A. (2014) Cathepsin S signals via PAR2 and generates a novel tethered ligand receptor agonist. PLoS One 9, e99702. 10.1371/journal.pone.0099702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R.; Mihara K.; Chung H.; Renaux B.; Lau C. S.; Muruve D. A.; DeFea K. A.; Bouvier M.; Hollenberg M. D. (2011) Neutrophil elastase acts as a biased agonist for proteinase-activated receptor-2 (PAR2). J. Biol. Chem. 286, 24638–24648. 10.1074/jbc.M110.201988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GraphPad Prism 7 for Windows, 7.04; GraphPad Software, Inc.: La Jolla, CA, 2017.
- PyMOL Molecular Graphics System, 2.0; Schrodinger, LLC: New York, 2017.
- Marvin 17.27.0; ChemAxon Ltd., 2017. http://www.chemaxon.com.
- Ballante F.; Marshall G. R. (2016) An Automated Strategy for Binding-Pose Selection and Docking Assessment in Structure-Based Drug Design. J. Chem. Inf. Model. 56, 54–72. 10.1021/acs.jcim.5b00603. [DOI] [PubMed] [Google Scholar]
- MacroModel, release 2016–3 v11.3; Schrödinger, LLC: New York, 2016.
- Maestro, release 2016–3; Schrödinger, LLC: New York, 2016.
- ROCS 3.2.0.4; OpenEye Scientific Software: Santa Fe, NM, 2013.
- Prime, release 2016–3; Schrödinger, LLC: New York, 2016.
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- BIOVIA Draw, 16.1.0.693; Dassault Systèmes: San Diego, CA, 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.