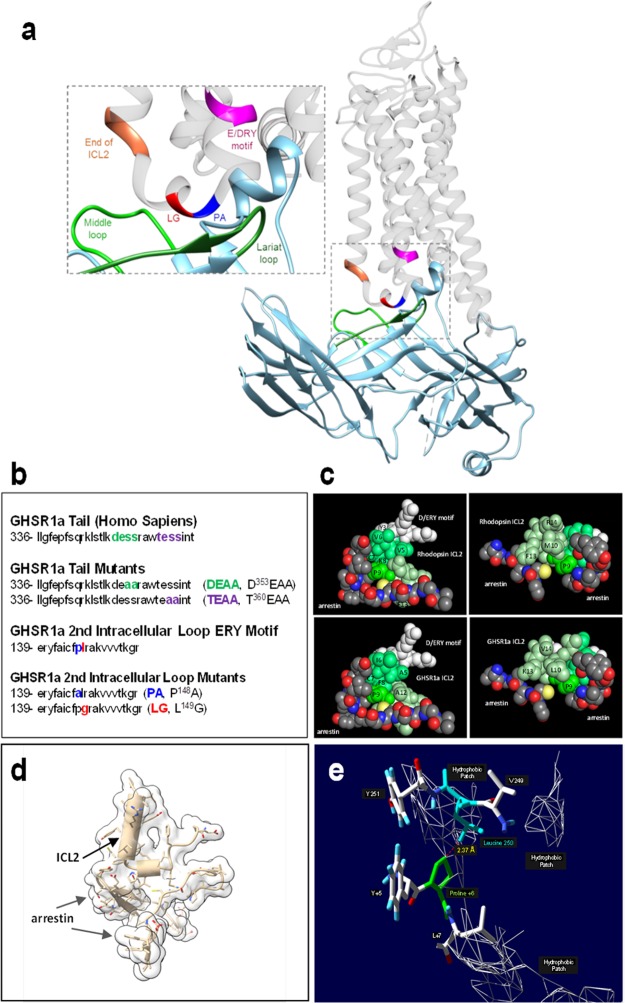
Figure 1.
Models of ICL2 arrestin structure for rhodopsin and GHSR1a substitution mutants. Graphics were generated from the crystal structure results of Kang et al.1 (a) Cartoon image of the visual-arrestin (in blue and green)/rhodopsin complex demonstrates the “core” conformation, in which both the receptor tail and the second intracellular loop (ICL2) are engaged. (b) Amino acid sequences of GHSR1a’s second intracellular loop (ICL2) and the C-tail. The D353ESS (DESS, green) and T360ESS (TESS, magenta) motifs in GHSR1a’s C-tail are putative regulatory sites for β-arrestin binding. In the ICL2 ERY motif, the +6 proline (PA, blue) and the +7 hydrophobic (leucine, LG, red) amino acid residues are putative β-arrestin and G-protein-binding sites, respectively. The two residues were mutated to alanine and glycine. (c) Complementary space filling model views of ICL2/arrestin engagement for rhodopsin (upper panels) and GHSR1a (lower panels). (d) Cartoon illustrating +6 proline N-terminal α helix capping of a short segment (smaller cylinder) of GHSR1a ICL2 in the active arrestin crevice. (e) Stick-figure image of the location of the GHSR1a (rhodopsin) ICL2+6 proline (green) relative to a C terminal arrestin middle loop hydrophobic patch containing leucine 250 (blue).