Fig. 6.
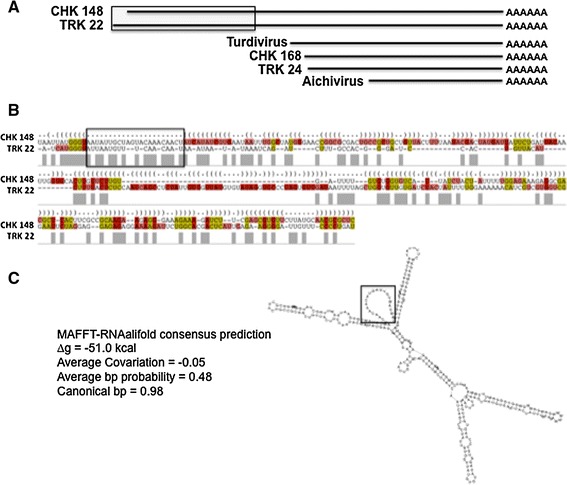
Secondary structural analysis of the CHK 148 and TRK 22 3′-NTRs. a Map of the region of the CHK 148 and TRK 22 3′-NTRs used for analysis. The gray box indicates the region aligned by secondary structure predictions in b and c. b The secondary structure alignment of the 5′-regions of the CHK 148 and TRK 22 3′-NTRs (250 nt). Structural alignments were generated using the IUPAC nucleotide ambiguity system. As in Fig. 5, the structure shown was predicted by MAFFT-RNAalifold program using the Webserver for Aligning non-structural RNAs (WAR, http://genome.ku.dk/resources/war/). c The predicted stem-loop structure (below right), and derived thermodynamic and statistical values for the proposed structure (∆g, avg covariation, avg bp probability, and canonical bp). Boxed sequences in the alignment in b correlate with the boxed loop in the secondary structure predication in c and is provided for reference and orientation
