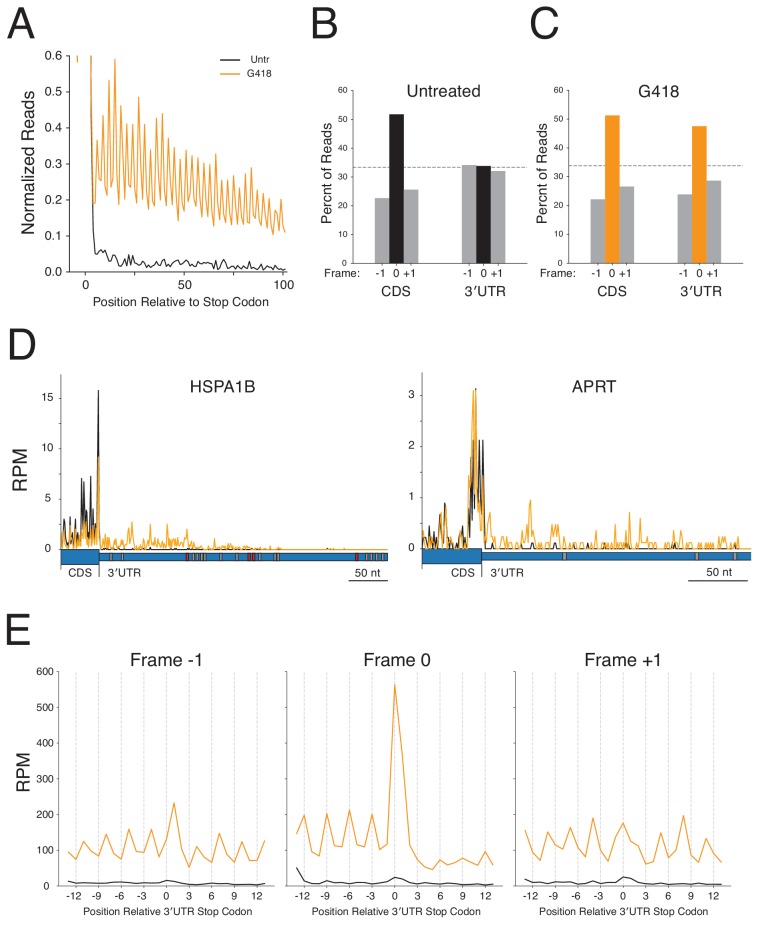
Figure 3. 3′UTR ribosomes in G418-treated cells derive from stop codon readthrough.
(A) Average gene plot showing increased density of ribosomes in 3′UTRs in G418-treated cells (orange) relative to untreated cells (black). Reading frame is analyzed for (B) Untreated and (C) G418-treated cells showing the percent of ribosomes in a given frame in the CDS and 3′UTR. (D) Gene models of HSPA1B (left) and APRT (right) showing translation of the 3′UTRs of these genes. G418-treated cells (orange lines) are overlaid onto untreated cells (black lines). The wider blue bar below the plot indicates the CDS and the narrow blue bar represents the 3′UTR. In-frame 3′TCs are colored in red, while out-of-frame 3′TCs are colored in gray. (E) Average gene plots show total ribosome density in the region surrounding the first in-frame 3′TC when found in frame −1 (left), frame 0 (center), or frame +1 (right). Transcripts with additional 3′TCs in this window were excluded for this analysis.