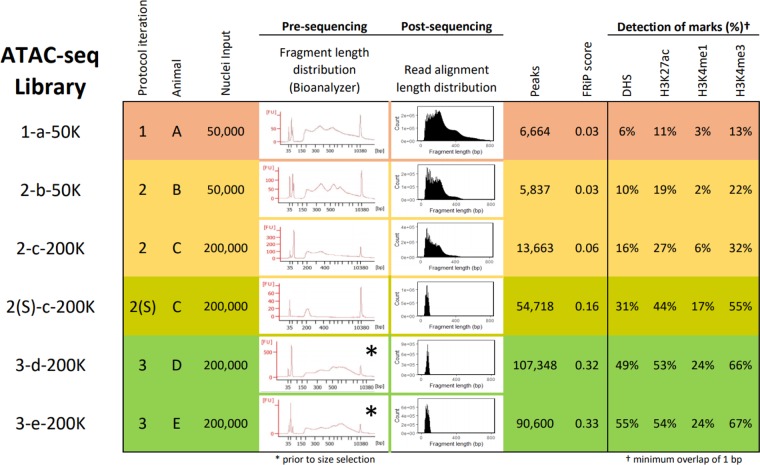
Figure 2.
Summary of representative libraries for each ATAC-seq protocol iteration, produced from cryopreserved chicken lung nuclei. Row colors indicate different iterations of the ATAC-seq protocol. Library names indicate “Protocol iteration–Animal–Nuclei input(–Replicate)”. Progressive changes to the protocol improved signal (Fraction of Reads in Peaks (FRiP) score) and improved overlap with DNase-I Hypersensitive Sites (DHS) and active histone modifications H3K27ac, H3K4me1, and H3K4me3 (minimum 1 bp overlap).