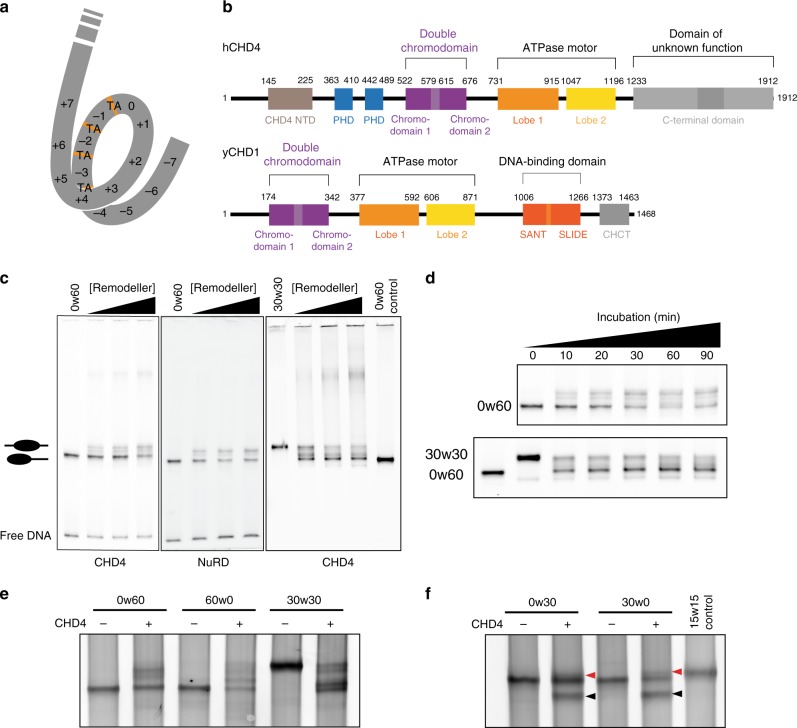
Fig. 1. CHD1/CHD4 domain topology and nucleosome repositioning assays.
a Schematic showing the 15 superhelical location (SHL) sites, the most outward-facing positions of the minor groove, in the 601 nucleosome positioning sequence (labelled +7 to –7). The histone–DNA interface consists mainly of inward-facing sections of the DNA minor groove, which are defined as superhelical locations (SHL) ± 0.5–6.531. Orange bands indicate four phased TpA dinucleotides that are spaced 10 bp apart. b Domain architectures of yeast CHD1 and human CHD4 with residues at domain boundaries indicated (NTD: N-terminal domain, PHD: plant homeodomain, CHCT: CHD1 C-terminal domain). c Gel-based nucleosome repositioning assays carried out with the indicated nucleosomes and remodellers. Fluorescently labelled nucleosomes were treated with the indicated remodeller for 60 min, the reaction was stopped by adding dsDNA (33 µg/mL) and then the samples were run on 5% native polyacrylamide gels. Symmetrically positioned nucleosomes are retarded relative to asymmetrically positioned species, as indicated. d Gel-based nucleosome repositioning assays carried out as described in (c), except that an incubation time-course was carried out at a single CHD4 concentration (5 nM). Assays using 0w60 (upper panel) or 30w30 (lower panel) substrates are shown; a 0w60 control lane was included in the lower panel. Source data are provided as a Source Data file. e, f Nucleosome sliding assays carried out as described in (c), using the indicated nucleosome substrates and 5 nM CHD4. Remodelled products (0w30 and 30w0) are indicated by red arrows, and the possible hexosome band is indicated by black arrows. Source data are provided as a Source Data file.