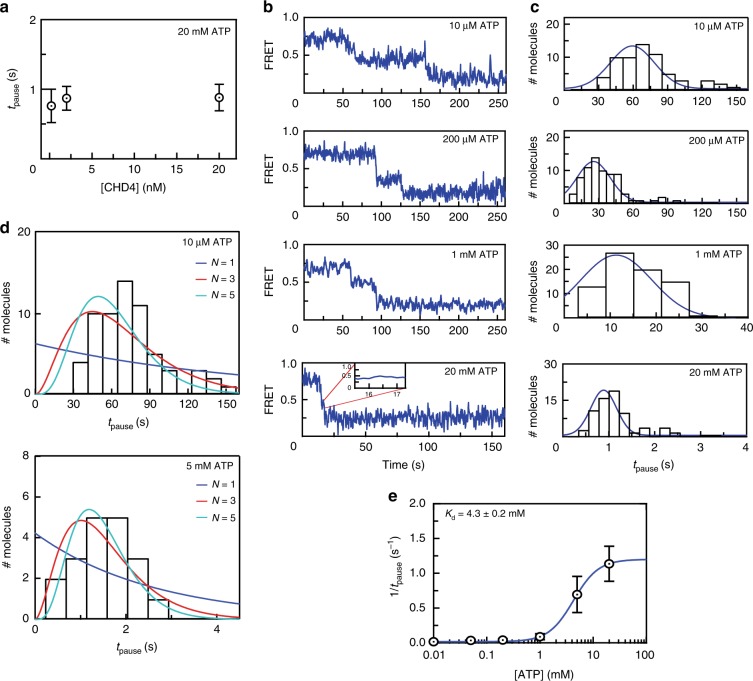
Fig. 4. CHD4-mediated nucleosome sliding is processive and depends on the binding of multiple ATP molecules.
a Distribution of tpause times for remodelling of 0AF555w60Bio at the indicated CHD4 concentrations and 20 mM ATP. Error bars represent standard deviation (of 45, 74 and 56 independent single particles at 0.2, 2 and 20 nM CHD4, respectively). b Typical traces from smFRET assays carried out with 2 nM CHD4 and at the indicated ATP concentrations. c Histograms showing tpause distributions of 70–82 particles from the experiments shown in (b). The histograms are fitted to a Gaussian distribution. d Pause time histograms for experiments carried out at 10 μM ATP (top) or 5 mM ATP (bottom) are overlaid with gamma distributions depicting different numbers of fundamental reaction steps (N = 1–5). e A 1:1 binding isotherm fit of the mean tpause time as a function of ATP concentration, with data taken from the assays in (c). Error bars represent standard deviation.