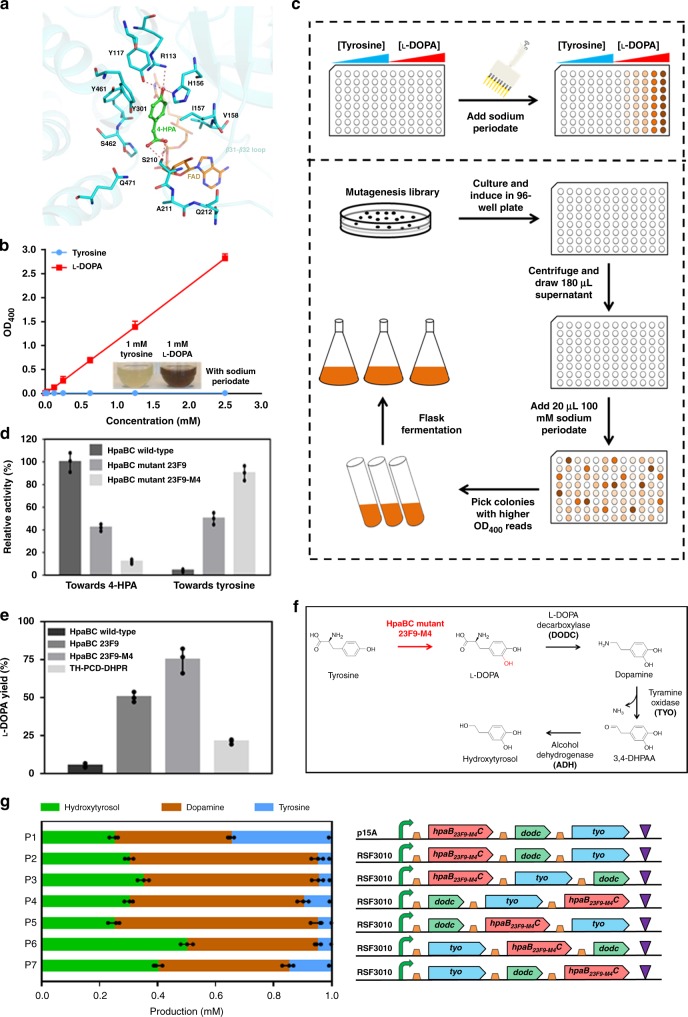
Fig. 1. Development of a microbial tyrosine hydroxylase and redesign of the hydroxytyrosol biosynthetic pathway.
a Model of HpaBC bound with 4-HPA and FAD. 4-HPA and FAD are shown as stick and colored in green and brown, respectively. Hydrogen bonds are shown as red dash. b The standard curve of the periodate assay. c Flowchart of the high-throughput screening of HpaBC. d Specific activities of the wild-type and mutant HpaBCs on 4-HPA and tyrosine. e l-DOPA yields of strain BW25113 expressing wild-type, HpaBC mutant 23F9, HpaBC mutant 23F9-M4 and the reported pathway enzymes TH-PCD-DHPR13. f Redesign of the hydroxytyrosol biosynthetic pathway using HpaBC mutant 23F9-M4 as a highly efficient tyrosine hydroxylase. g Optimization of plasmid copy number and gene orders in the operon encoding the biosynthetic pathway. The concentration of the indicated compounds was determined, and no l-DOPA was accumulated (The green, brown and blue bars indicate the concentrations of hydroxytyrosol, dopamine and tyrosine, respectively). 4-HPA, 4-hydroxyphenylacetate; 3,4-DHPAA, 3,4-dihydroxyphenyl acetaldehyde. The data shown in b, d, e and g are from three replicate experiments and are expressed as the mean±SD. Source data underlying Figs. 1b, d, e, and g are provided as a Source Data file.