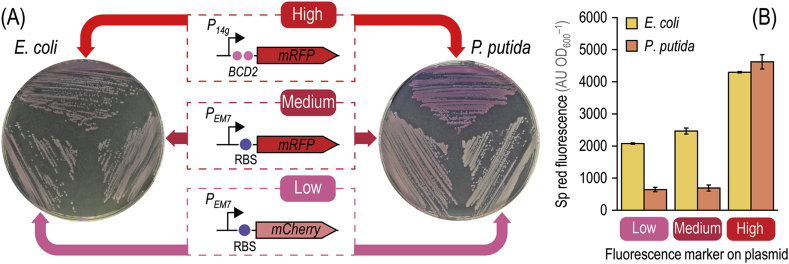
Fig. 2.
Insertion of red fluorescent modules into plasmids carrying the meganuclease gene. (A) Plasmid pSEVA228S (XylS/Pm→I-SceI, KmR) was used as a template to generate a family of derivatives carrying fluorescent modules yielding low, medium, and high levels of red fluorescence. The modules contain the genes encoding either mCherry or the monomeric red fluorescent protein (mRFP) under transcriptional control of the synthetic, constitutive PEM7 or P14g promoters. Each gene is preceded by a regulatory element, indicated by a purple circle, composed of a ribosome binding site and a short spacer sequence (5′-AGG AGG AAA AAC AT-3′). For the module yielding high levels of red fluorescence, a bicistronic design (BCD2) was used as a translational coupler. E. coli DH5α and P. putida KT2440 were transformed with derivatives of plasmid pSEVA228S (Table 1) and streaked onto LB medium plates containing Km. E. coli colonies were incubated at 37 °C for 18 h and photographed afterwards, while P. putida colonies were grown at 30 °C for 18 h and plates were stored at 8 °C for a further 24 h to allow for fluorophore maturation. (B) Specific (Sp) red fluorescence in cultures of E. coli DH5α and P. putida KT2440 transformed with plasmid pS228SR·L (low), pS228SR·M (medium), or pS228SR (high). Cells were grown in LB medium added with Km for 18 h and the Sp red fluorescence, expressed as arbitrary units (AU) relative to the optical density measured at 600 nm (OD600) of the cultures, was measured after resuspending the bacterial pellets in M9 minimal medium. Each bar represents the mean value of the Sp red fluorescence in each culture ± standard deviation of quadruplicate measurements from at least three independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)