Fig. 2.
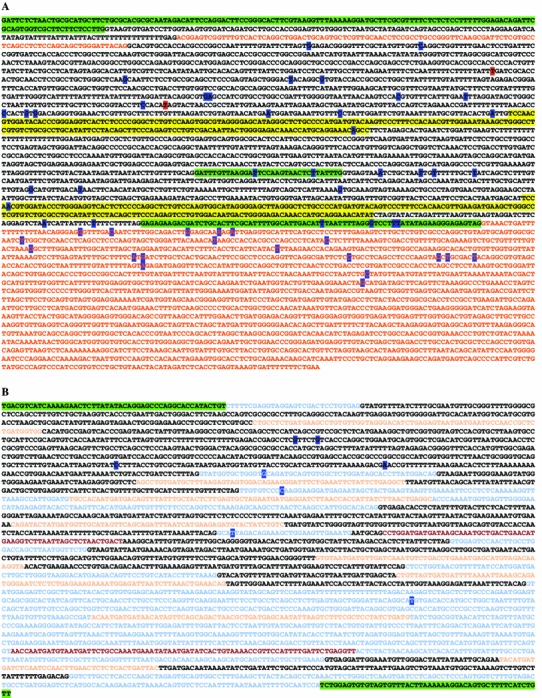
a Gene disruptions within the RCC1, SNHG3, SNORA73A, and SNORA73B loci observed in clonal cell lines resisting lytic viral infection. The SNHG3 gene is a non-coding transcript that shares exons with the protein-coding RCC1 gene (highlighted in green). Introns are designated with black text, whereas SNHG3 exonic sequences not shared with RCC1 are shown in red. SNORA73A and SNORA73B are shown as the first and second intronic sequences highlighted in yellow, respectively. Gene-trap insertions were observed within introns, as well each of the above mentioned genes. Insertion sites are shown with single letters highlighted in blue (human cell lines resisting DFV2, FLU, HRV2, or HRV16 infection) or red (Vero E6 monkey cells resisting HSV2 infection). b SNHG2 and intervening SNORD sequences. Gene-trap insertion sites conferring resistance to pathogens are highlighted in blue text. In some cases, identical clones were recovered from viral selection in independent studies with more than one virus (viruses shown in Table 1). Color coding in 5′ to 3′ orientation: maroon—SNORD44 and SNORD47; light orange—SNORDs 74–81; light blue—SNHG2 coding sequence; black—SNHG2 introns; green—adjacent 5′ and 3′ genomic sequence. Three SNORDs are within coding the sequence for GAS 5, namely SNORDs 47, 76, and 80 (Color figure online)
