Figure 6.
Simulations of TAC-Based Membrane Tubulation and Comparison of In Silico and Experimental Data
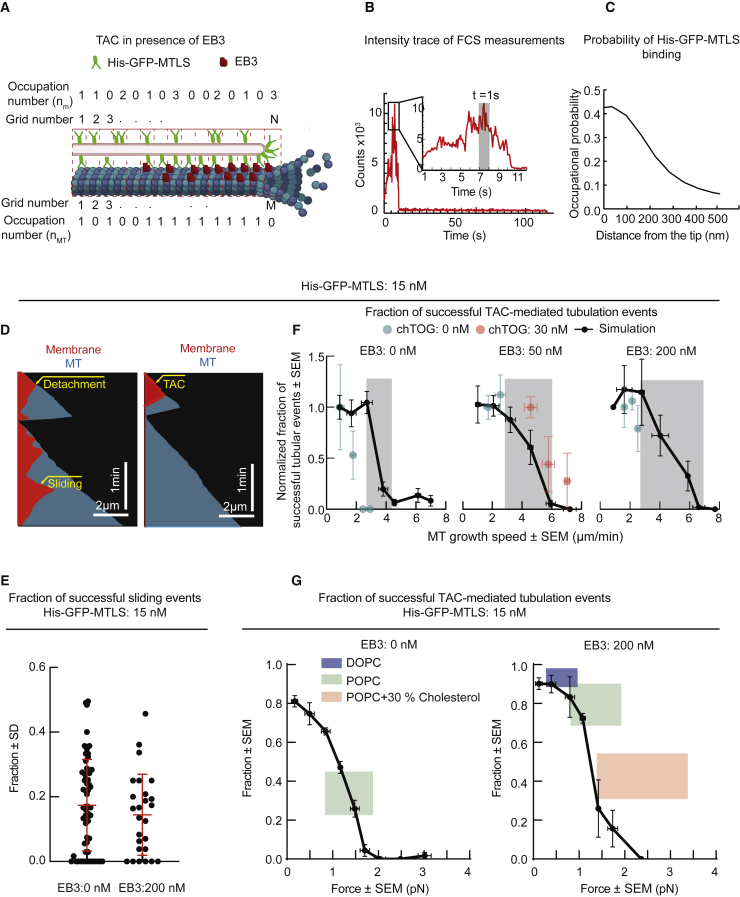
(A) Kinetic scheme for the simulations of a membrane sliding at the MT tip.
(B) Representative intensity time trace of a MT tip labeled with EB3-GFP recorded by FCS. The inset shows maximum intensity during a time window of 12 s. The gray rectangle marks the intensity in 1 s.
(C) Estimated occupational probability of His-GFP-MTLS at the MT tip in the presence of 200 nM EB3.
(D) Kymographs obtained from the stochastic simulations of membrane spreading in the presence of dynamic MTs.
(E) Fraction of successful sliding-driven tubulation events, calculated from simulations in the same way as in Figure 3H. n, number of independent simulations (black dots) EB3 0 nM, 58 (1,007 traces); EB3 200 nM, 28 (348) traces.
(F) Fraction of membrane deformations becoming tubular as a function of MT growth rate in simulations and experiments. Data were normalized to the average fraction obtained at the lowest MT growth rate. Gray rectangles mark the areas where the probability decays with the speed. n, number of events experiments: 0 nM EB3, 50 (7); 200 nM EB3, 68 (9); 50 nM EB3, 28 (7); 30 nM chTOG; and 50 nM EB3, 22 (4). Simulations: EB3 0 nM, 17 (87 traces); EB3 50 nM, 57 (1510 traces); EB3 200 nM, 28 (348 traces). Every experimental point represents the mean speed, calculated from several individual experiments inside intervals of 0.5 μm/s.
(G) Fraction of membrane deformations becoming tubular as a function of the force needed to pull a tube. n, number of experiments: POPC, EB3 0 nM and His-GFP-MTLS 15 nM, 48 (7); POPC, EB3 200 nM and His-GFP-MTLS 15 nM, 68 (9); POPC+30% cholesterol, EB3 200 nM and His-GFP-MTLS 15 nM, 61 (3); DOPC, EB3 200 nM and His-GFP-MTLS 15 nM, 22 (2). Simulations: EB3 0 nM, 59 (824 traces); EB3 200 nM, 31 (424 traces). The experimentally measured values are represented by rectangles due to the experimental uncertainty.
See also Table S1.