Abstract
Background:
Endogenous phospholipase A2 inhibitors from snake blood (sbPLIs) have been isolated from several species around the world, with the primary function of self-protection against the action of toxic phospholipases A2. In American snakes, sbPLIs were solely described in pit vipers, in which the natural protection role is justified. In this study, we described a sbPLI in Boa constrictor (popularly known as jiboia), a non-venomous snake species from America.
Methods:
PLA2 inhibitory activity was tested in the blood plasma of B. constrictor using C. d. terrificus venom as the enzyme source. Antibodies developed against CNF, a sbγPLI from Crotalus durissus terrificus, were used to investigate the presence of homologues in the blood plasma of B. constrictor. A CNF-like molecule with a PLA2 inhibitory activity was purified by column chromatography. The encoding gene for the inhibitor was cloned from B. constrictor liver tissue. The DNA fragment was cloned, purified and sequenced. The deduced primary sequence of interest was aligned with known sbγPLIs from the literature.
Results:
The blood plasma of B. constrictor displayed PLA2 inhibitory activity. A CNF-like molecule (named BcNF) was identified and purified from the blood plasma of B. constrictor. Basic properties such as molecular mass, composing amino acids, and pI were comparable, but BcNF displayed reduced specific activity in PLA2 inhibition. BcNF showed highest identity scores (ISs) with sbγPLIs from pit vipers from Latin America (90-100%), followed by gamma inhibitors from Asian viperid (80-90%). ISs below 70% were obtained for BcNF and non-venomous species from Asia.
Conclusion:
A functional sbγPLI (BcNF) was described in the blood plasma of B. constrictor. BcNF displayed higher primary identity with sbγPLIs from Viperidae than to sbγPLIs from non-venomous species from Asia. The physiological role played by sbγPLIs in non-venomous snake species remains to be understood. Further investigation is needed.
Keywords: Phospholipase A2 inhibitor, Phospholipase A2, Boidae, Snakes, Rattlesnake venom
Background
Secretory phospholipases A2 are widely distributed as toxic components of snake venoms. A number of snake species express endogenous snake blood phospholipase A2 inhibitors (sbPLIs). This kind of molecules was first described in venomous snakes with the primary function of self-protection against an eventual presence of snake venom PLA2 (svPLA2) in their own blood stream [1, 2]. According to the presence of known domains from mammal proteins - C-type lectin-like, tandem leucine-rich repeats (LRRs), or three-finger motifs - sbPLIs were grouped into alpha (α), beta (β) or gamma (γ) structural classes, respectively [3]. Comparable inhibitors were later identified in a number of non-venomous species [4, 5, 6, 7, 8, 9]. Whether venomous or not, some snake species express sbPLIs belonging to up to three different structural classes simultaneously [3, 7, 10, 11].
SbγPLIs are the most widely distributed inhibitors among elapid and viperid species from the Old and New World [12, 13, 14]. Concerning non-venomous snakes, as far as we know, until now sbγPLIs were solely purified from Asian species [4, 5, 6, 7, 8, 9]. With that in mind, we investigated the presence of this kind of inhibitor in Boa constrictor - a non-venomous tropical snake - popularly known as jiboia. We identified a functional sbγPLI, cloned the encoding gene from liver tissue and structurally characterized the deduced protein. The sbγPLI was named BcNF by analogy with CNF (Crotalus neutralizing factor), a prototype of this class of inhibitors previously isolated from the South American rattlesnake, Crotalus durissus terrificus [15, 16].
Methods
Boa constrictor blood plasma and liver tissue collection
Heparinized blood plasma and liver tissue fragments were collected from a Boa constrictor specimen captured in the municipality of Contagem (19º55'54" S, 44º03'13" W), in the Brazilian state of Minas Gerais. The specimen was kept in captivity in the Serpentarium of Ezequiel Dias Foundation until death by natural causes. The whole blood was collected immediately after the animal death, centrifuged for plasma separation and clarified using a 0.22-µm microfilter. The total protein content was estimated by spectrophotometry readings at 280 nm. One optical density unit was considered to be equivalent to 1 mg/mL of protein. Liver fragments were collected in DEPC-treated tubes and quickly frozen in liquid nitrogen. Whenever applicable, blood plasma and tissue liver from C. d. terrificus specimens were used as reference.
Fractionation of B. constrictor blood plasma
Five hundred microliters of B. constrictor blood plasma were diluted to 10 mL with 25 mM Tris-HCl, 0.1 M NaCl pH 8.7 (buffer A) and dialyzed against the same buffer to ensure ionic equilibrium. After centrifugation to remove any insoluble material, the supernatant was loaded into an anion exchange column (Hitrap QFF 1mL, GE HealthCare). Protein elution was performed with a linear gradient of 25 mM Tris-HCl, pH 8.7, containing 2.0 M NaCl (buffer B), under a flow rate of 1 mL/min. Fractions with inhibitory activity (1 mL each) were pooled, 4-fold diluted with a saturated ammonium sulfate (SAS) solution and loaded into hydrophobic interaction columns connected in series [four columns HiTrap Phenyl FF 5 mL (low sub) column, GE HealthCare]. Elution was performed with a decreasing salt gradient under a flow of 5 mL/min. Total protein concentration was estimated by optical density readings of the eluted fractions at 280 nm.
Inhibition of PLA2 activity
The crude venom of C. d. terrificus was used as a source of PLA2. Increasing volumes of snake blood plasma with known protein concentration were preincubated with a fixed concentration (50 μg/mL) of C. d. terrificus venom for 30 min at 37°C. The same procedure was applied to purified fractions, after dialysis against 25 mM ammonium formats, pH 6.5, whenever necessary. Residual PLA2 activity was evaluated by measuring the clearing halos (in mm) of hydrolysis in agar gels with incorporated hen egg yolk suspension [17]. Negative (PBS) and positive (no blood plasma) controls were run in parallel. Inhibition curves were constructed by plotting the halo diameter against protein concentration in logarithm scale. Data were analyzed by linear regression using least squares method in the Graph Prism 6.0 for Mac OS X (GraphPad software Inc., California). Curve limits were calculated with 95% of confidence level. Specific activities were represented by curve slopes and expressed by mean ± S.D. Whenever applicable, regression line slopes were statistically compared in pairs.
SDS-PAGE and western blotting
B. constrictor blood plasma and purified BcNF were analyzed by SDS-PAGE in a 15% homogeneous or in an 8-25% gradient Phast® gel (Phast System®, GE HealthCare). Western blotting was revealed with rabbit anti-CNF IgG (0.5 mg/mL), followed by commercial anti-rabbit IgG-peroxidase antibody (A0545, Sigma) at a 1:5000 dilution. The color reaction was developed with DAB (3,3' diaminobenzidine tetrahydrochloride) in the presence of H2O2.
RNA extraction and cDNA synthesis
Total RNA was isolated from about 50 mg of B. constrictor liver tissue using Trizol® (Invitrogen, USA) following the manufacturer’s instructions. RNA integrity was analyzed by gel electrophoresis in a 0.8% agarose gel using TBE (89 mM Tris base, 89 mM boric acid, 2 mM EDTA, pH 8.0) as running buffer. RNA bands were visualized under UV light, after staining with ethidium bromide. After cDNA synthesis using 2 to 5 (g of total RNA and oligo(dT)12-18 primer (First-Strand Synthesis kit, Invitrogen, USA), polymerase chain reactions were carried out with specific oligonucleotides based on the primary structure of CNF [15]: 3’CGCTCATGTGACTTTTGTCAC5’ (sense, amino-terminus), 3’TCAGAGGCTTGCCAATCTGATG5’ (antisense, carboxy-terminus). A housekeeping gene (β?actin) was PCR-amplified in parallel, in the presence of adequate oligonucleotides.
Fresh PCR products were cloned into the pGEM-T vector (Promega, USA) following the manufacturer’s instructions. Insert-containing clones were isolated after PCR screening of transformed NM522 E. coli. Negative control contained no DNA. Amplified products were analyzed by electrophoresis in 1.0 % agarose gels in TBE buffer, in the presence of ethidium bromide. DNA from three positive clones were completely sequenced by the dideoxy chain termination method [18] on an automated ABI Prism 310 Genetic Analyzer (Perkin Elmer Applied Biosystems, USA) with the Big Dye Terminator Cycle Sequencing Ready Reaction (Perkin Elmer Applied Biosystems, USA). M13 forward and M13 reverse oligonucleotides were used as primers.
The cycling conditions were 3 min at 94°C, 35 cycles of 30 sec at 94°C, 30 sec at 55°C and 1 min at 72°C, followed by an extension period of 5 min at 72°C in a TC412 thermocycler (Techne).
Primary/secondary structure predictions and multiple alignment
Three complete reads in both directions were assembled and aligned against each other. The consensus sequence was used to deduce the primary structure and main basic properties of BcNF. The secondary structure was predicted using the Chou Fasman algorithm. Multiple sequencing alignments with primary structures of other sbγPLIs were performed using the ClustalW algorithm and a Gonnet’s similarity matrix was subsequently generated. Inclusion criterium for sbγPLIs was the access to chemically determined or deduced primary structures in public data bases. For species with two or more sequence deposits due to isoforms, calculated consensus was taken as representative of the inhibitor. Signal peptides were removed, whenever necessary. The sbγPLIs from the following snake species were aligned: Bothrops alternatus (ABV91326/7), Bothrops erythromelas (ABV91328/9), Bothrops jararaca (ABV91330/1), Bothrops jararacussu (ABV91332/3), Bothrops moojeni (ABV91332/5), Bothrops neuwiedi (ABV91336/7), Crotalus durissus terrificus (AAA19162), Elaphe climacophora (BAH47550), Elaphe quadrivirgata (BAA83078), Gloydius brevicaudus (formerly Agkistrodon blomhofii siniticus) (BAA86970), Lachesis muta (AAR04437/8), Malayopython reticulatus (formerly Python reticulatus) (AAF73945), Notechis scutatus (CAB56615/6/7), Oxyuranus microlepidotus (AAF23784), Oxyuranus scutellatus (AAF23781), Protobothrops flavoviridis (formerly Trimeresurus flavoviridis) (BAA24502), Protobothrops elegans (BAJ14719/20/21), Pseudonaja textilis (AAF23783), Sinonatrix annularis (JN975878). All the procedures were performed using the MacVector 16.0.10 software (Mac Vector Inc., USA) with default parameters.
Results
Identification and purification of BcNF from B. constrictor blood plasma
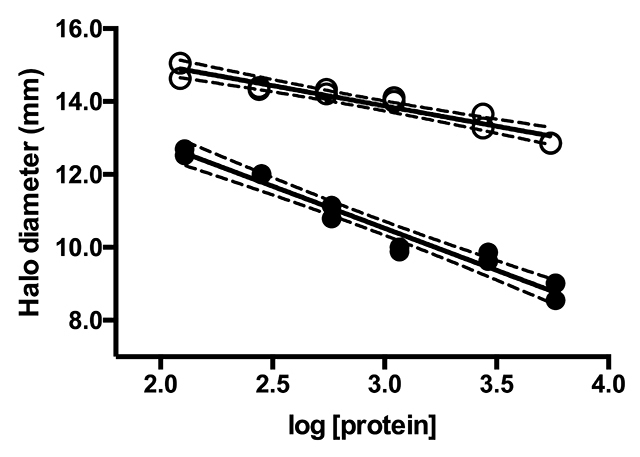
First, the blood plasma of B. constrictor was tested for inhibition of C. d. terrificus venom PLA2 (Fig. 1). Inhibition was observed, although to a lesser extent when compared to C. d. terrificus blood plasma (positive control). Specific activities for PLA2 inhibition were: -1.112 ± 0.1075 and -2.307 ± 0.1498 for B. constrictor and C. d. terrificus blood plasma, respectively. These activities were statistically different (p < 0.0001). PLA2 inhibition activity was significantly lower for B. constrictor blood plasma.
Figure 1. Inhibition curves of PLA2 activity of C. d. terrificus venom by the blood plasma of Boa constrictor (white dots). The blood plasma of C. d. terrificus was used as reference (black dots). Curve equations: y = (-1.112 ± 0.1075)x + (17.22 ± 0.3193) for B. constrictor and y = (-2.307 ± 0.1498)x + (17.45 ± 0.4478) for C. d. terrificus, with determination coefficient (R2) of 0.9145 and 0.9546, respectively. The 95% confidence intervals of the best fit curves are indicated by dashed lines.
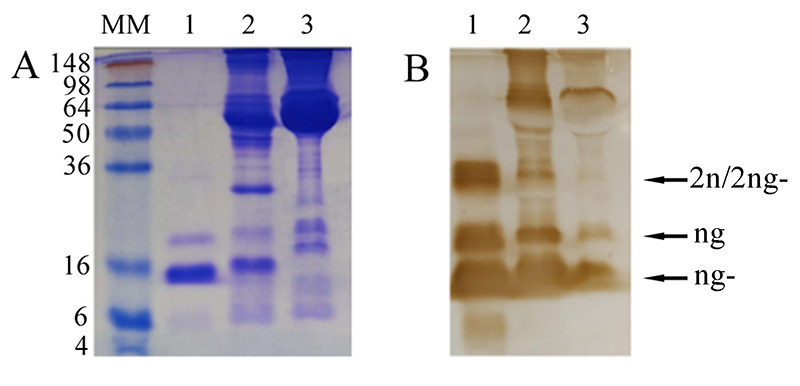
Following, we investigated whether the observed inhibition could be due to the presence of a sbγPLIs. Western blotting revealed the presence of a CNF-like molecule in the blood plasma of B. constrictor. Two main protein bands were recognized by anti-CNF antibodies (Fig. 2), with apparent molecular masses roughly corresponding to glycosylated (ng) and non-glycosylated (ng-) monomers. A fainter band was present with mol. mass of possible dimers (2ng/2ng-). The result indicated the presence of a sbγPLIs, named BcNF, in the blood plasma of B. constrictor.
Figure 2. (A) SDS-PAGE 15% after staining with Coomassie Blue and (B) Western blotting revealed with anti-CNF IgG. Lanes: MM - molecular marker (in kDa) (SeeBlue Plus 2 Pre-stained Protein Standard, Invitrogen); 1 - CNF (20 µg); 2 - C. d. terrificus blood plasma (80 µg); 3 - B. constrictor blood plasma (80 µg). The arrows indicate non-glycosylated monomer (ng-), glycosylated monomer (ng) and possible dimers (2ng/2ng-).
BcNF was isolated from B. constrictor blood plasma using two chromatographic steps: an ionic exchange followed by a hydrophobic interaction. The eluted fractions were assayed for PLA2 inhibition (Fig. 3). Fractions from the second purification step presenting inhibitory activity were combined and submitted to electrophoresis and Western blotting using anti-CNF IgG. A CNF-like molecule (BcNF) was mostly eluted with 100% of ultrapure water (Fig. 4). BcNF and CNF (positive control) at varying concentrations were assayed for PLA2 inhibition (Fig. 5). Calculated specific activities were -1.344 ± 0.1705 and -4.797 ± 0.3434 for BcNF and CNF, respectively. These activities were statistically different (p < 0.0001). BcNF inhibited PLA2 at a significant lesser extent compared to CNF.
Figure 3. Purification of BcNF from B. constrictor blood plasma. (A) Anion-exchange and (B) hydrophobic interaction chromatograms. Elution gradients are indicated by dotted lines. PLA2 inhibitor-containing fractions are indicated by horizontal bars.
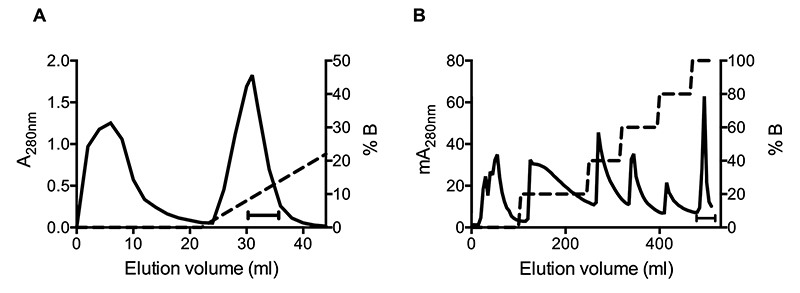
Figure 4. (A) SDS-PAGE in 8-25% gel after silver staining and (B) Western blotting developed with anti-CNF IgG. PC: positive control (CNF). Lanes are numbered on top according to percentages of eluent B (ultrapure water) in the hydrophobic interaction chromatography.

Figure 5. Inhibition curves of PLA2 activity of C. d. terrificus venom by BcNF isolated from B. constrictor blood plasma (white dots). CNF from C. d. terrificus snakes was used as positive control for PLA2 inhibition (black dots). Curve equations: y = (-1.344 ± 0.1705)x + (13.50 ± 0.4235) for BcNF, and y = (-4.797 ± 0.3434)x + (19.13 ± 0.4478) for CNF, with determination coefficient (R2) of 0.8860 and 0.9606, respectively. The 95% confidence intervals of the best fit curves are indicated by dashed lines.

BcNF cloning from B. constrictor liver tissue
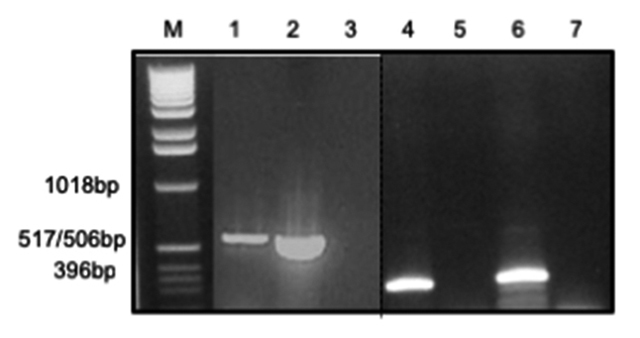
The integrity of extracted RNA from B. constrictor liver tissue was confirmed by the unique presence of characteristic bands corresponding to 18S and 28S ribosomal RNAs (data not shown). After RT-PCR in the presence of specific primers for CNF, an amplicon of about 545 bp confirmed the encoding of a CNF-like protein in the liver tissue of B. constrictor (Fig. 6). The DNA fragment was cloned, purified and sequenced for further analysis.
Figure 6. Electrophoresis of RT-PCR products after amplification of liver tissue with specific primers for CNF (left side) or β-actin (right side). M: molecular marker 1 kb DNA ladder (Gibco-BRL). Lanes 1 and 4: B. constrictor liver; lanes 2 and 6: C. d. terrificus liver (reference); lanes 3 and 7: negative control (no DNA); lane 5: no reverse transcriptase in the reaction.
Deduced primary structure and chemical properties predictions of BcNF
The deduced primary sequence of mature BcNF was compared to that of CNF. Both proteins are composed of 181 amino acids, including 16 conserved cysteines and a single putative N-linked carbohydrate site at Asn157. Fourteen amino acid substitutions were noted in BcNF when compared to CNF, one of them (R93/K93) within a segment proposed before for sbγPLIs interaction with PLA2 (Fig. 7). Basic properties of BcNF and CNF are summarized in Table 1.
Figure 7. Alignment of the deduced primary structure of BcNF (sbγPLI from B. constrictor) and CNF (sbγPLI from C. d. terrificus). Identical amino acids are in grey background. Amino acid substitutions are in white background. The decapentapeptide Q84PFPGLPLSRPNGYY98 is indicated by a continuous black arrow above the numbering line.
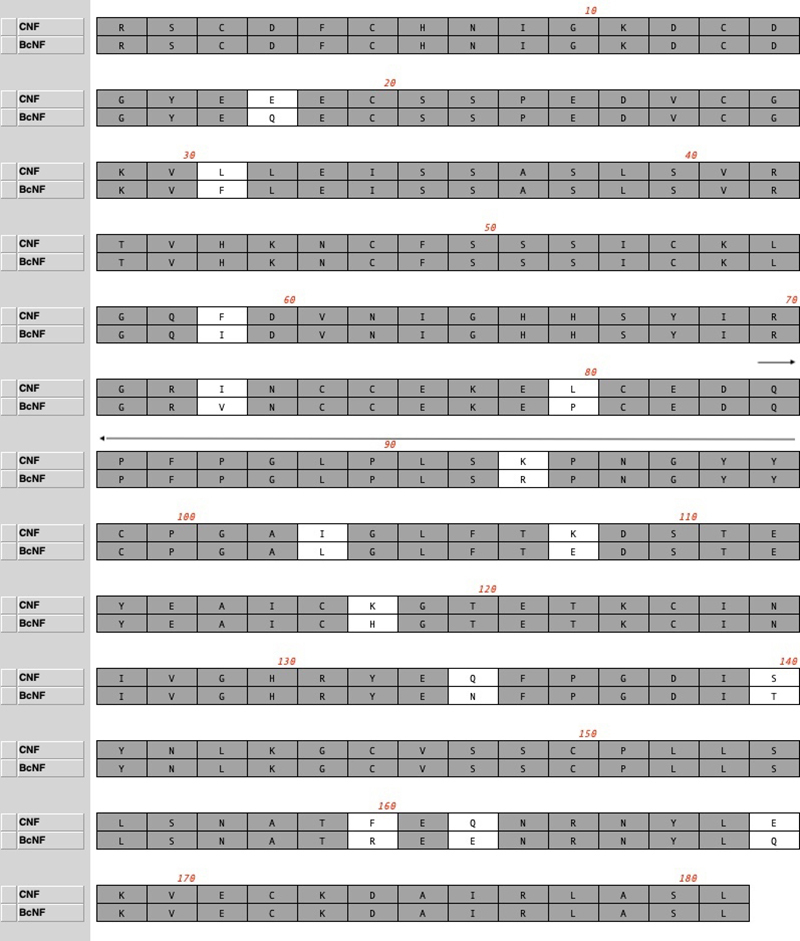
Table 1. Comparison of basic properties of BcNF (sbγPLI from B. constrictor) and CNF (sbγPLI from C. d. terrificus).
| Property | BcNF | CNF |
|---|---|---|
| Molecular mass (Da) | 20074.57 | 20058.69 |
| Isoelectric point (pI) | 5.51 | 5.55 |
| Composing amino acids | ||
| Total (no.) | 181 | 181 |
| Chemical character (%) | ||
| Non-polar | 30.4 | 30.9 |
| Polar | 43.0 | 42.4 |
| Acidic | 13.2 | 13.3 |
| Basic | 13.3 | 13.3 |
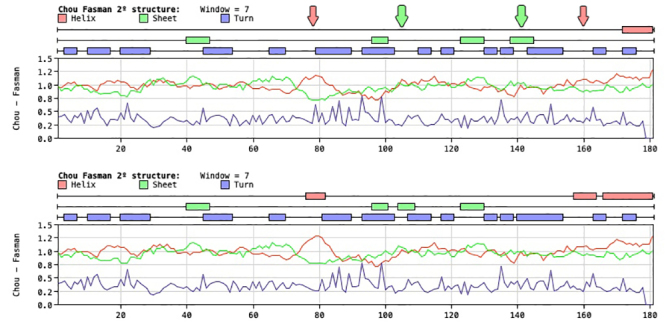
Amino acid substitutions, in general, lead to a decrease in the number of α-helixes from three in CNF to one in BcNF, besides a displacement of beta sheets in the predicted secondary structures of the proteins (Fig. 8).
Figure 8. Secondary structure predicted for BcNF (top) compared to CNF (bottom). The differences are indicated by arrows on top of BcNF structure, using the same color as in the structural diagram.
Multiple sequence alignment of BcNF and other sbγPLIs
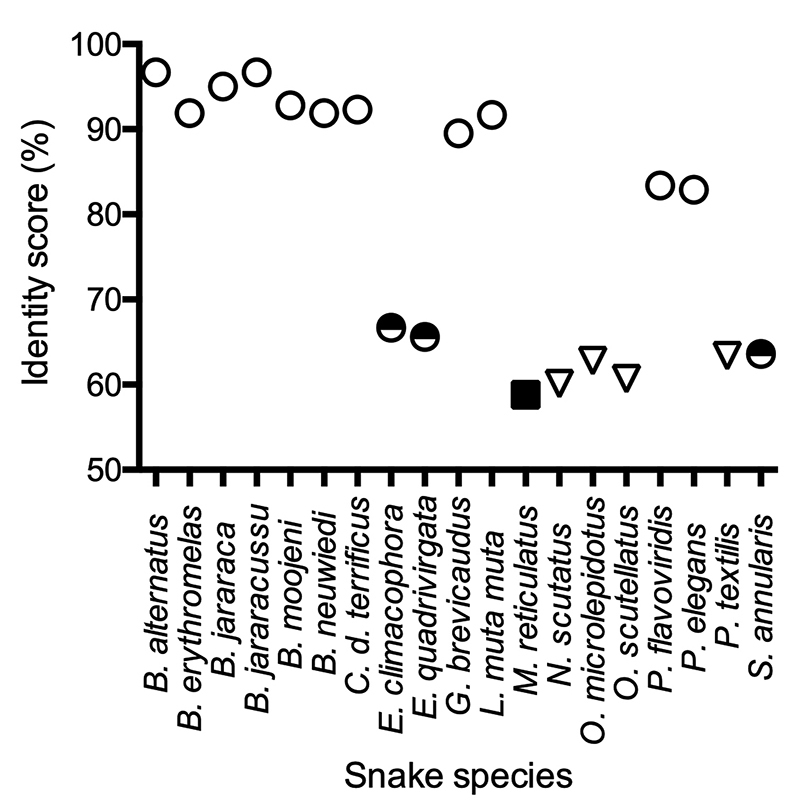
The deduced primary sequence of BcNF was multiply aligned with sbγPLIs from venomous and nonvenomous snakes from Asia, Australia and Latin America (available as Additional file 1). A similarity matrix was generated (available as Additional file 2) and the identity scores (ISs) were graphically represented (Fig. 9). For BcNF and sbγPLIs from Latin American pit vipers, most ISs were within the last decile (90-100%). ISs above 80% were obtained for Asian viperid snakes. On the other hand, when BcNF was compared to sbγPLIs from non-venomous species from Asia, the ISs were below 70%. ISs below 70% were also obtained for Elapidae snakes.
Figure 9. Graphical representation of the identity scores (ISs) obtained in Gonnet’s similarity matrix after multiple alignment of the deduced primary structure of BcNF with known sbγPLIs. Black/white circle: Colubridae, white triangle: Elapidae, white circle: Viperidae, black rectangle: Pythonidae.
Discussion
Boa is a Neotropical genus of snakes that occurs almost continuously from southern South America through to northern Mexico [19]. Historically recognized as monotypic, recent data based on the distinct morphological traits, color patterns exhibited by these snakes and the wide diversity of ecosystems they inhabit, collectively suggest that the genus contains multiple species [20]. In Brazil, B. constrictor (sensu lato) can be found all over the country, except in the extreme south [21]. It is an aglyphous species, devoid of venom or Duvernoy’s glands. Similarly to other henophidian snakes (boas, pythons and their kin), B. constrictor uses constriction to subdue and kill a wide range of prey - including lizards, birds and mammals - through an interesting modulated process mediated by the victim’s heartbeat [22]. Apparently, there is no need of an inhibitor for self-protection against toxic svPLA2.
The detection of sbPLIs in non-venomous snake species is not a novelty. The first sbγPLIs was isolated from E. quadrivirgata [6]. The finding was later attributed to feeding habits of the species on venomous snakes [7]. However, another sbγPLI - named PIP for phospholipase A2 inhibitor from Python - was soon described in the non-venomous and non-ophiophagus species Malayopython reticulatus (formerly Python reticulatus) [8]. Since then, a number of sbγPLIs were detected in colubrid from Asia: Dinodon rufozanatum [5], Elaphe carinata [5], E. climacophora [7], E. rufodorsata [5], E. teniura, Macropisthodon rudis [9], Synonatrix annularis [4], and Zaocys dhumnades [5], in addition to xenodermatid Achalinus rufescens [5]. A structurally-related PIP homolog was also described in the non-venomous rock python (P. sebae) from Africa, although with poor PLA2 inhibition activity [23]. Regarding non-venomous snakes living in the American continent, studies are lacking on any sbPLI.
B. constrictor inhibition of PLA2 was lower than that of C. d terrificus blood plasma. Similarly, BcNF was less active than CNF. Our results are in accordance with those described for E. climacophora and E. quadrivirgata. Respective sbγPLIs were detected at higher amounts in the former, and justified by the ophiophagous habits of the species [7]. It is important to note that, in addition to sbγPLIs, those Elaphe species express sbα? and sbβPLIs simultaneously in the circulating blood. We used antibodies developed against CNF to search for sbγPLI in B. constrictor. The detection of inhibitors from other structural classes is a possibility that cannot be discarded.
BcNF is highly similar to CNF, with 14 substitutions in a total of 181 amino acids and an IS of about 90%. The molecular masses of the non-glycosylated monomers, calculated from amino acid compositions, are very close (Table 1). Band migrations in gel electrophoresis also indicated similar apparent molecular masses for monomers and oligomers (Fig. 7). Like CNF, BcNF is composed by a mixture of non-glycosylated (20 kDa) and glycosylated (22-24 kDa) monomers. For CNF, which is the main subject of study in our lab, the proportion between non-glycosylated and glycosylated varies according to the preparation. The sample loaded in SDS-PAGE (Fig. 2) was mostly non-glycosylated. However, it has been shown that the carbohydrate moiety is not essential for PLA2 inhibition by CNF [24]. The same might be true for BcNF. The tendency for oligomerization might be a shared property, too. In fact, the 16th, 113th, 132nd and 166th tyrosinyl residues, which were previously suggested to form the interface between monomers in the oligomerization of CNF, are maintained at the same positions in BcNF. These residues might be involved in the oligomerization of the latter also. BcNF was only tested against svPLA2 from C. d. terrificus venom, but the possibility of inhibition of other svPLA2 cannot be discarded. The decapentapeptide Q84PFPGLPLSRPNGYY98, which was previously proposed to be the best consensus motif possibly involved in the sbγPLIs interaction with PLA2s is maintained in BcNF. The only amino acid replacement was conservative (R93/K93).
Interestingly, BcNF appeared more closely related to sbγPLIs from Latin American pit vipers, and from Asian pit vipers to a lesser extent, than to those from non-venomous snakes from Asia described so far.
Conclusion
A functional sbγPLI (BcNF) was described, for the first time, in the blood plasma of B. constrictor, a non-venomous species from America. BcNF displayed higher primary identity with sbγPLIs from pit vipers than with sbγPLIs from non-venomous species from Asia. Even with a growing number of sbγPLI identifications in the last years, the physiological role played by these proteins in non-venomous snake species remains to be clarified.
Abbreviations
IS: identity score; LRRs: tandem leucine-rich repeats; SAS: saturated ammonium sulfate; sbPLI: snake blood phospholipase A2 inhibitor; svPLA2: snake venom phospholipase A2.
Acknowledgments
The authors would like to thank A. C. Valentim, L. A. Melo, R. M. Lima, P. S. Almeida for the technical support and the undergraduate students V. L. R Perché and M. P. Romualdo for their contribution.
Supplementary material
The following online material is available for this article:
Footnotes
Availability of data and materials: All data extracted and or analyzed during this study are included in this published article.
Funding: C. L. Fortes-Dias is a productivity fellow from the State of Minas Gerais Research Foundation (FAPEMIG), a Brazilian funding agency.
Ethics approval: Not applicable.
Consent for publication: Not applicable.
References
- Kihara H. Studies on phospholipase A in Trimeresurus flavoviridis venom. III. Purification and some properties of phospholipase A inhibitor in Habu serum. J Biochem. 1976;80(2):341–349. doi: 10.1093/oxfordjournals.jbchem.a131282. [DOI] [PubMed] [Google Scholar]
- Ovadia M, Kochva E, Moav B. The neutralization mechanism of Vipera palaestinae neurotoxin by a purified factor from homologous serum. Biochim Biophys Acta. 1977;491(2):370–386. doi: 10.1016/0005-2795(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Ohkura N, Okuhara H, Inoue S, Ikeda K, Hayashi K. Purification and characterization of three distinct types of phospholipase A2 inhibitors from the blood plasma of the Chinese mamushi, Agkistrodon blomhoffii siniticus. Biochem J. 1997;325(2):527–531. doi: 10.1042/bj3250527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Z, Li X, Yuan P, Liu P, Huang C. Orthogonal optimization of prokaryotic expression of a natural snake venom phospholipase A2 inhibitor from Sinonatrix annularis. Toxicon. 2015;108:264–271. doi: 10.1016/j.toxicon.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Li J, Xiong Y, Sun S, Yu L, Huang C. Preparation of monoclonal antibodies against gamma-type phospholipase A2 inhibitors and immunodetection of these proteins in snake blood. J Venom Anim Toxins incl Trop Dis. 2017:23–37. doi: 10.1186/s40409-017-0128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K, Masui K, Inoue S, Ikeda K, Hayashi K. Purification, characterization and cDNA cloning of a phospholipase A2 inhibitor from the serum of the non-venomous snake Elaphe quadrivirgata. Biochem J. 1999;341(1):165–171. [PMC free article] [PubMed] [Google Scholar]
- Shirai R, Toriba M, Hayashi K, Ikeda K, Inoue S. Identification and characterization of phospholipase A2 inhibitors from the serum of the Japanese rat snake, Elaphe climacophora. Toxicon. 2009;53(6):685–692. doi: 10.1016/j.toxicon.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Twin MM, Gopalakrishnakone P, Kini RM, Armugam A, Jeyaseelan K. Recombinant antitoxic and anti-inflammatory factor from the nonvenomous snake Python reticulatus: phospholipase A2 inhibitor and venom neutralizing potential. Biochemistry. 2000;39(31):9604–9611. doi: 10.1021/bi000395z. [DOI] [PubMed] [Google Scholar]
- Zhong L, Huang C. Isolation and biochemical characterization of a gamma-type phospholipase A2 inhibitor from Macropisthodon rudis snake serum. Toxicon. 2016;122:1–6. doi: 10.1016/j.toxicon.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Nobuhisa I, Chiwata T, Fukumaki Y, Hattori S, Shimohigashi Y, Ohno M. Structural elements of Trimeresurus flavoviridis serum inhibitors for recognition of its venom phospholipase A2 isozymes. FEBS Lett. 1998;429(3):385–389. doi: 10.1016/s0014-5793(98)00602-4. [DOI] [PubMed] [Google Scholar]
- Lizano S, Angulo Y, Lomonte B, Fox JW, Lambeau G, Lazdunski M, et al. Two phospholipase A2 inhibitors from the plasma of Cerrophidion (Bothrops) godmani which selectively inhibit two different group-II phospholipase A2 myotoxins from its own venom: isolation, molecular cloning and biological properties. Biochem J. 2000;346:631–639. [PMC free article] [PubMed] [Google Scholar]
- Campos PC, de Melo LA, Dias GLF, Fortes-Dias CL. Endogenous phospholipase A2 inhibitors in snakes: a brief overview. J Venom Anim Toxins incl Trop Dis. 2016;22(37) doi: 10.1186/s40409-016-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli CG, Borges RJ, Fernandes CAH, Matioli FM, Fernandes CFC, Conceição J, Sobrinho, et al. Molecular cloning and structural modelling of gamma-phospholipase A2 inhibitors from Bothrops atrox and Micrurus lemniscatus snakes. Int J Biol Macromol. 2017;103:525–532. doi: 10.1016/j.ijbiomac.2017.05.076. [DOI] [PubMed] [Google Scholar]
- Serino-Silva C, Morais-Zani K, Hikari Toyama M, Toyama DO, Gaeta HH, Rodrigues CFB, et al. Purification and characterization of the first γ-phospholipase inhibitor (γPLI) from Bothrops jararaca snake serum. PLoS One. 2018;13(3):e0193105. doi: 10.1371/journal.pone.0193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes-Dias CL, Lin Y, Ewell J, Diniz CR, Liu TY. A phospholipase A2 inhibitor from the plasma of the South American rattlesnake (Crotalus durissus terrificus). Protein structure, genomic structure, and mechanism of action. J Biol Chem. 1994;269(22):15646–15651. [PubMed] [Google Scholar]
- Perales J, Villela C, Domont GB, Choumet V, Saliou B, Moussatché H, et al. Molecular structure and mechanism of action of the crotoxin inhibitor from Crotalus durissus terrificus serum. Eur J Biochem. 1995;227(1-2):19–26. doi: 10.1111/j.1432-1033.1995.tb20355.x. [DOI] [PubMed] [Google Scholar]
- Habermann E, Hardt KL. A sensitive and specific plate test for the quantitation of phospholipases. Anal Biochem. 1972;50(1):163–173. doi: 10.1016/0003-2697(72)90495-2. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron RA, Reynolds RG, Burbrink FT. A taxonomic revision of Boas (Serpentes: Boidae) Zootaxa. 2014;3846(2):249–260. doi: 10.11646/zootaxa.3846.2.5. [DOI] [PubMed] [Google Scholar]
- Card DC, Schield DR, Adams RH, Corbin AB, Perry BW, Andrew AL, et al. Phylogeographic and population genetic analyses reveal multiple species of Boa and independent origins of insular dwarfism. Mol Phylogenet Evol. 2016;102:104–116. doi: 10.1016/j.ympev.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puorto G, França FOSF. Serpentes não peçonhentas. Aspectos clínicos dos acidentes. In: Cardoso JLC, editor. Animais peçonhentos no Brasil: biologia, clínica e terapêutica dos acidentes. Ed. Sarvier; São Paulo: 2009. pp. 108–114. Venomous animals in Brazil: biology, clinical and therapeutic accidents. [Google Scholar]
- Boback SM, Hall AE, McCann KJ, Hayes AW, Forrester JS, Zwemer CF. Snake modulates constriction in response to prey's heartbeat. Biol Lett. 2012;8(3):473–476. doi: 10.1098/rsbl.2011.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnini S, Finetti F, Francese S, Boscaro F, Dani FR, Maset F, et al. A novel protein from the serum of Python sebae, structurally homologous with type-γ phospholipase A2 inhibitor, displays antitumour activity. Biochem J. 2011;440(2):251–262. doi: 10.1042/BJ20100739. [DOI] [PubMed] [Google Scholar]
- Fortes-Dias CL, Ortolani PL, Fernandes CA, Lobo KR, Amaral de Melo L, Borges MH, et al. Insights on the structure of native CNF, an endogenous phospholipase A2 inhibitor from Crotalus durissus terrificus, the South American rattlesnake. Biochim Biophys Acta. 2014;1844(9):1569–1579. doi: 10.1016/j.bbapap.2014.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


