Figure 4. Loss of Dscam1-L Impairs Axon Projection.
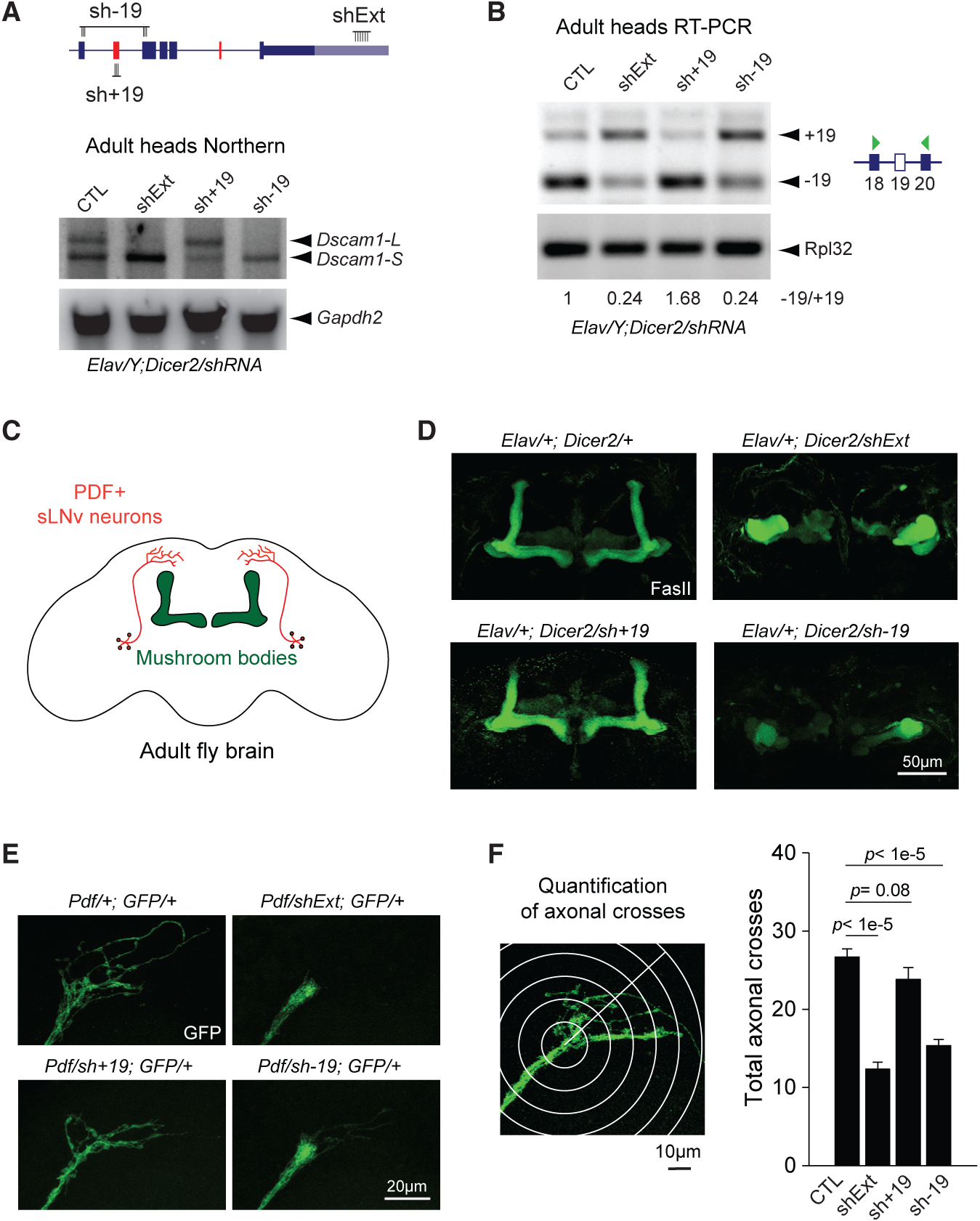
(A) Top: schematic of shRNA constructs targeting specific Dscam1 splice junctions (sh−19, sh+19) and the long 3′ UTR (shExt). Bottom: northern blot of male heads from Elav-driven shRNA knockdown shows loss of Dscam1-L upon knockdown with sh−19 or shExt, but not sh+19. Gapdh2 northern is shown as a loading control.
(B) RT-PCR shows exon 19 skipping patterns under knockdown conditions in adult heads.
(C) Schematic showing central brain and MB anatomy, with MB highlighted in green and sLNv neurons highlighted in red.
(D) Staining of adult brains with anti-Fasciclin II (FasII) reveals massive disorganization of the central brain and impaired bifurcation of the MBs upon neuronal knockdown using shExt and sh−19, but not sh+19.
(E) Impaired axonal outgrowth in adult sLNv neurons expressing sh−19 or shExt driven by Pdf-Gal4.
(F) Left: methodology of axonal outgrowth quantification of PDF-positive sLNv neurons. Right: Sholl analysis demonstrates that both shExt and sh−19 flies have significantly reduced axonal crossing compared with controls. Error bars represent SEM. n = 16–19.
Elav represents Elav-GAL4, Dicer2 represents UAS-Dicer2, shExt represents UAS-shExt, sh−19 represents UAS-sh−19, sh+19 represents UAS sh+ 19, and GFP represents UAS-mCD8::EGFP.
