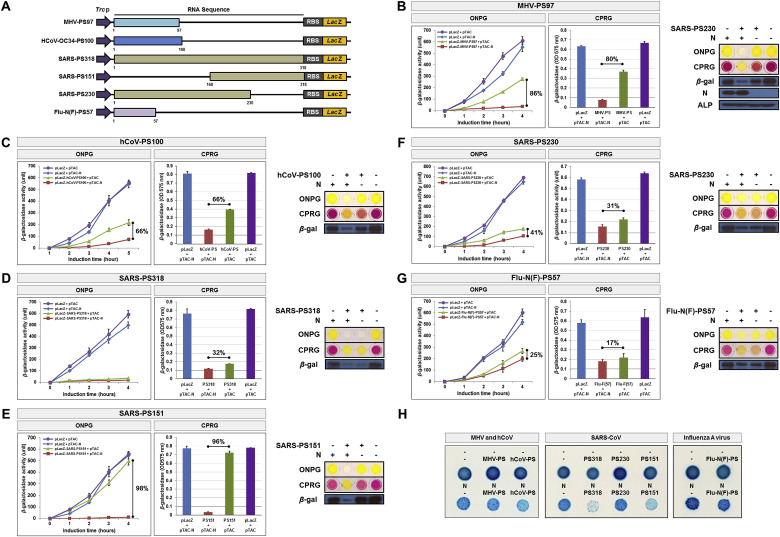
Fig. 2.
The N-protein specifically recognizes viral RNA packaging signals from the coronaviridae family. (A) Schematic representation of the packaging signal (PS) sequence regions of coronaviruses (mouse hepatitis virus, human coronavirus, SARS-CoV) and the influenza A virus, as well as the deletion constructs used to probe regions necessary for N-protein binding. Numbers in parenthesis indicate nucleotide lengths of the sequences designated. (B) Effect of N-protein on mouse hepatitis virus PS-lacZ reporter gene expression, and various regions of the PS-RNA sequence (C–G). β-galactosidase activities achieved upon transformation with individual reporter constructs or control vectors, examined in the presence (red; square) and absence (green; triangle) of N-protein, respectively. pLacZ and pTAC are control plasmids that lack expression of either the packaging signal sequence or N-protein, respectively. Results of the ONPG and CPRG assays measuring the interaction of the SARS-CoV N-protein and each packaging signal RNA are shown on the right, as well as a Western blot demonstrating the expression of LacZ (β-galactosidase) at 4 h after IPTG induction. Alkaline phosphatase (ALP) was probed as a loading control. All experiments were performed in triplicate, and data are presented as the mean ± SD (standard deviation) of three separate experiments. (H) Assay for LacZ translation on X-gal/IPTG agar plates upon co-transformation of JM109 cells with SARS-CoV N-protein and CoV packaging sequences. Minus (-) and N show JM109 transformants containing CoV packaging signal RNAs in the absence and presence of N-protein, respectively. All experiments were performed three times. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)