Fig. 1.
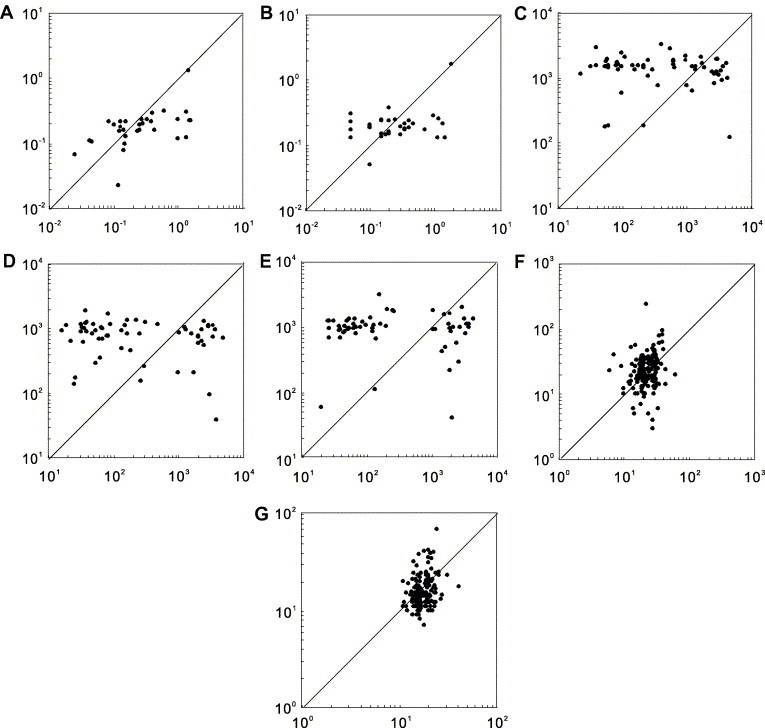
Comparisons of mRNA stabilities between intronless genes and intron-containing genes with similar mRNA lengths and functions. The X axis represents intronless genes, while the Y axis shows their intron-containing counterpart genes. The distribution of dots below and above the right angle bisector intuitively illustrates the comparisons of mRNA stability between intronless genes and intron-containing genes. Meanwhile, we performed Wilcoxon signed ranks test to determine the significance of the difference. (A) mRNA decay rate (h−1) in human HepG2 cells, 31 gene pairs, P = 4.77 × 10−3; (B) mRNA decay rate (h−1) in human Bud8 cells, 34 gene pairs, P = 0.033; (C) mRNA half-life (min) in human T lymphocytes (with medium), 62 gene pairs, P = 0.043; (D) mRNA half-life (min) in human T lymphocytes (with an Anti-CD3), 62 gene pairs, P = 0.666; (E) mRNA half-life (min) in human T lymphocytes (with anti-CD3 and anti-CD28), 62 gene pairs, P = 0.375; (F) mRNA half-life (min) in Saccharomyces cerevisiae strain Y262, 205 gene pairs, P = 3.04 × 10−4; and (G) mRNA half-life (min) in S. cerevisiae strain rpb1-1, 201 gene pairs, P = 3.88 × 10−9.
