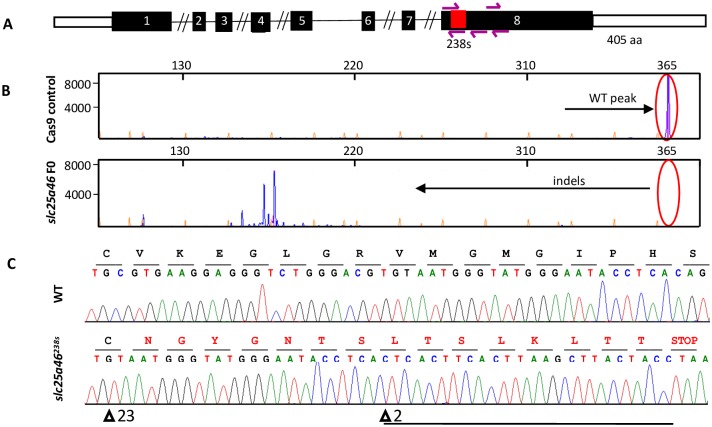
Fig 1.
Genome editing of slc25a46 gene in zebrafish using CRISPR/Cas9 system: (A) Schematic diagram of zebrafish slc25a46 gene with a frameshift in exon 8 indicated as a red box and a premature stop codon at amino acid position 238 (allele slc25a46238s). The magenta arrows indicate five CRISPR guides injected together to create both F0 crispants and stable mutants. (B) Fragment analysis of the amplified CRISPR targeted region from the FAM-labeled PCR product (365 base pair amplicon) showing WT peak in control and multiple peaks indicating insertions and deletions (indels) in a representative slc25a46 F0 sample. X-axis represents base pair number; Y-axis represents signal intensity. (C) Sequence traces of wild-type (WT) and slc25a46238s stable mutant zebrafish: exon 8, residues 223–238 indicating homozygous frameshift (in red); deletions are indicated with a triangle, insertion is underlined.