The complex bacterial populations that constitute the gut microbiota can harbor antibiotic resistance genes (ARGs), including those encoding β-lactamase enzymes (BLA), which degrade commonly prescribed antibiotics such as ampicillin. The prevalence of such genes in commensal bacteria has been increased in recent years by the wide use of antibiotics in human populations and in livestock. While transfer of ARGs between bacterial species has well-established dramatic public health implications, these genes can also function in trans within bacterial consortia, where antibiotic-resistant bacteria can provide antibiotic-sensitive neighbors with leaky protection from drugs, as shown both in vitro and in vivo, in models of lung and subcutaneous coinfection.
KEYWORDS: antibiotic resistance, gut microbiota, infection
ABSTRACT
The complex bacterial populations that constitute the gut microbiota can harbor antibiotic resistance genes (ARGs), including those encoding β-lactamase enzymes (BLA), which degrade commonly prescribed antibiotics such as ampicillin. The prevalence of such genes in commensal bacteria has been increased in recent years by the wide use of antibiotics in human populations and in livestock. While transfer of ARGs between bacterial species has well-established dramatic public health implications, these genes can also function in trans within bacterial consortia, where antibiotic-resistant bacteria can provide antibiotic-sensitive neighbors with leaky protection from drugs, as shown both in vitro and in vivo, in models of lung and subcutaneous coinfection. However, whether the expression of ARGs by harmless commensal bacterial species can destroy antibiotics in the intestinal lumen and shield antibiotic-sensitive pathogens is unknown. To address this question, we colonized germfree or wild-type mice with a model intestinal commensal strain of Escherichia coli that produces either functional or defective BLA. Mice were subsequently infected with Listeria monocytogenes or Clostridioides difficile, followed by treatment with oral ampicillin. The production of functional BLA by commensal E. coli markedly reduced clearance of these pathogens and enhanced systemic dissemination during ampicillin treatment. Pathogen resistance was independent of ARG acquisition via horizontal gene transfer but instead relied on antibiotic degradation in the intestinal lumen by BLA. We conclude that commensal bacteria that have acquired ARGs can mediate shielding of pathogens from the bactericidal effects of antibiotics.
INTRODUCTION
Antibiotic administration has markedly reduced the morbidity and mortality associated with bacterial infections in the preantibiotic era. Increasing antibiotic-resistance in pathogenic microbes, mediated in part by acquired genes that encode antibiotic-degrading enzymes, represents a major threat to human health (1).
The gut microbiota contains trillions of commensal bacteria that can also harbor antibiotic resistance genes (ARGs) (2). Notably, antibiotic exposure can increase ARG gene representation and expression by the gut microbiota (3). Horizontal ARG transfer represents a mechanism by which drug-sensitive microbes can acquire resistance, e.g., by acquisition of genes encoding antibiotic-degrading hydrolases (4, 5). Thus, it is possible that commensal bacterial species transfer ARGs to intestinal pathogens upon antibiotic exposure in the gut lumen. However, another possibility is that production of antibiotic-degrading enzymes by the resident intestinal microbiota protects otherwise drug-sensitive pathogens in trans, thereby facilitating their replication and spread in the host. In fact, within microbial communities leaky protection from β-lactams and chloramphenicol can be provided to antibiotic-sensitive bacteria by antibiotic-degrading microbes, as shown both in vitro and in vivo, using mouse models of pneumonia and subcutaneous abscesses (6–12). Importantly, antibiotic degradation could be operated by bacteria belonging to the Enterobacteriaceae family (11) or Bacteroides genus (10, 12), which are highly represented in the human and mouse intestinal tract, suggesting that pathogen-shielding antibiotic degradation might occur also in the gut. Based on these considerations, we set out to investigate whether harmless autochthonous bacteria might degrade orally administered antibiotics in the intestinal lumen, thereby impairing their ability to combat intestinal pathogens.
RESULTS
To test this hypothesis in a controlled system, we reconstituted germfree (GF) mice with an E. coli strain, utilized here as a model commensal, that expresses either a wild-type (WT) form of β-lactamase (TEM-1) or an inactive point mutant (here referred to as WT β-lactamase enzyme [BLA] or mut BLA, respectively) (Fig. 1A) (13). This approach yielded cohorts of mice that, with the exception of one codon, harbor identical metagenomes, thus excluding differences in microbiota functions (e.g., immune activation, colonization resistance, etc.) that are not related to the β-lactam degradation.
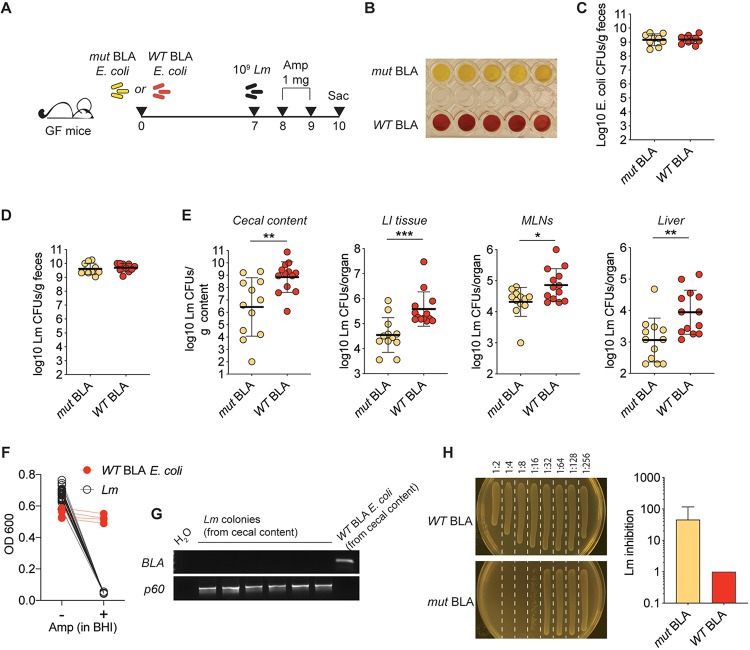
FIG 1.
β-Lactamase production by a model commensal curtails the efficacy of ampicillin against L. monocytogenes. (A) Schematic representation of the experimental design. (B) A nitrocefin assay was performed on resuspended fecal pellets obtained from the depicted groups of mice. Each well represents a different mouse. The results of one representative experiment of three performed are shown. (C) Reconstitution levels for the depicted E. coli strains, as measured by plating of fecal pellets on day 7 after reconstitution (day of infection), onto selective plates (n = 8 to 10; data are pooled from two independent experiments; shown are individual data points and geometric means). (D) Luminal L. monocytogenes burdens in the depicted mice at 1 day postinfection, measured by plating fecal pellets onto selective plates. (E) L. monocytogenes burdens in the depicted compartments at day 3 postinfection. In panels D and E, data are pooled from three independent experiments (n = 12) and shown as individual data points and geometric means (Mann-Whitney test: *, P < 0.05; **, P < 0.01; ***, P < 0.001). (F) Individual L. monocytogenes colonies (n = 28) or WT BLA E. coli colonies (n = 4) from four to five different mice were inoculated into BHI with or without ampicillin. The OD was measured after overnight culture. (G) Colonies utilized for the experiment depicted in panel F were also subjected to PCR with primers specific for the TME-1 β-lactamase gene or p60 (L. monocytogenes positive control). Shown are results for six L. monocytogenes colonies and one E. coli colony. Identical results were obtained for all tested colonies. (H) The cecal contents of WT mice reconstituted with either WT or mut BLA E. coli and administered ampicillin in drinking water for 2 days were serially diluted and inoculated with L. monocytogenes. L. monocytogenes growth was assessed after overnight culture by measuring the OD and direct plating (one representative plate per condition shown on the left). Plotted values correspond to the first dilution allowing for detectable L. monocytogenes growth (with 1 indicating L. monocytogenes growth at all dilutions; n = 3, means ± standard deviations are shown). Similar results were obtained utilizing antibiotic-treated, E. coli-reconstituted animals (see Fig. S1 in the supplemental material).
Although WT BLA and mut BLA E. coli reached identical luminal bacterial densities in reconstituted mice, a colorimetric assay confirmed that only the intestinal content of mice reconstituted with WT BLA E. coli retained the capacity to hydrolyze β-lactams (Fig. 1B and C). At 1 week after reconstitution, mice were orally infected with the foodborne pathogen Listeria monocytogenes 10403s. L. monocytogenes is highly sensitive to β-lactam antibiotics and can expand in the gut lumens of mice that lack colonization resistance (14). Mice were then administered ampicillin on days +1 and +2 after L. monocytogenes infection and sacrificed on day +3. As expected, L. monocytogenes reached identical densities in the intestines of WT BLA or mut BLA E. coli reconstituted mice on day +1, indicating that the two E. coli strains did not differ in their inability to provide colonization resistance against L. monocytogenes (Fig. 1D). However, we found significantly higher L. monocytogenes burdens in multiple organs in mice harboring WT BLA E. coli on day +3, consistent with the notion that β-lactamase-dependent ampicillin degradation shielded L. monocytogenes from the therapeutic antibiotic’s action (Fig. 1E).
To exclude the possibility that L. monocytogenes might have acquired resistance to ampicillin via horizontal gene transfer, we inoculated single L. monocytogenes colonies recovered from the cecal contents of WT BLA E. coli-reconstituted, L. monocytogenes-infected mice into liquid medium either in the presence or in the absence of ampicillin. Notably, none of the inoculated L. monocytogenes colonies grew in the presence of ampicillin, in contrast to WT BLA-expressing E. coli colonies recovered from the same mice (Fig. 1F). Furthermore, none of the L. monocytogenes colonies tested positive for the presence of the β-lactamase gene, which was uniformly detected in colonies of WT BLA E. coli by PCR (Fig. 1G).
To confirm that the increased L. monocytogenes burden observed above was due to antibiotic degradation by resident E. coli, we collected the cecal contents of mice reconstituted with either WT BLA or mut BLA E. coli and treated with ampicillin in the drinking water for two consecutive days to allow for luminal accumulation of the antibiotic. Inoculation of L. monocytogenes into serial dilutions of the cecal content supernatants revealed that the cecal contents from GF mice reconstituted with mut BLA E. coli had a higher inhibitory capacity than cecal contents recovered from mice reconstituted with WT BLA E. coli (Fig. 1H). These results were replicated using WT mice reconstituted with the model E. coli strains following antibiotic treatment (see Fig. S1 in the supplemental material). Since the presence of active β-lactamase was the only bona fide difference between the cecal contents of the two cohorts of mice used in the above-described experiments, we conclude that the microbiota-encoded enzymatic activity curtailed the efficacy of ampicillin treatment against L. monocytogenes.
To expand our observations beyond the Listeria model and to assess whether commensal-mediated antibiotic degradation may represent a mechanism that is relevant to other infectious agents, we adapted our experimental strategy to an established infection model using C. difficile (15) (Fig. 2A), an important intestinal pathogen that is also sensitive to ampicillin (Fig. S2). Of note, this model allowed us to investigate the relevance of our findings in a setting where expansion of an antibiotic-resistant microbe takes place following antibiotic-mediated depletion of the intestinal microbiota, a common occurrence in hospitalized patients (16). Importantly, expansion of the utilized E. coli strains in this setting was restrained by competition with the residual autochthonous flora (Fig. 2B), which allowed us to verify the effects of BLA-mediated ampicillin degradation in a context of bona fide reduced enzymatic concentration.
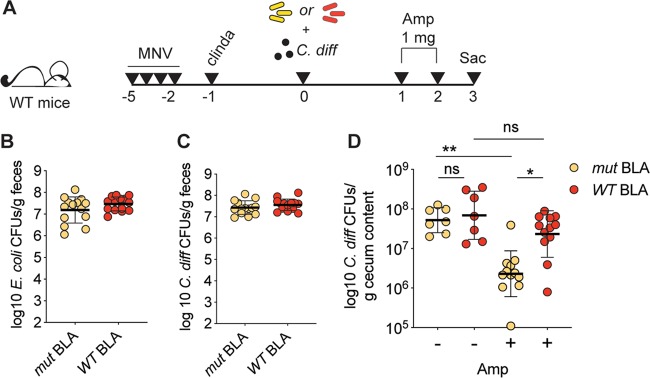
FIG 2.
Endogenous antibiotic degradation impacts treatment of C. difficile infection. (A) Schematic representation of the experimental design. (B) Reconstitution levels for mice reconstituted with either WT or mut BLA E. coli (as depicted in panel A) at 1 day after oral gavage, as assessed by selective plating of fecal pellets (n = 14; data are pooled from two independent experiments; individual data points and geometric means are shown). (C) C. difficile burdens in mice treated as depicted in panel A at 1 day after oral gavage, as assessed by selective plating of fecal pellets (n = 14; data are pooled from two independent experiments; individual data points and geometric means are shown). (D) C. difficile burden in the cecal content of mice treated as depicted in panel A at day 3 postinfection (n = 7 for controls; n = 12 for ampicillin treated). Data are pooled from three independent experiments. Individual data points and geometric means are shown (Kruskal-Wallis test with multiple comparisons: *, P < 0.05; **, P < 0.01).
Similar to the results obtained with L. monocytogenes, we observed indistinguishable levels of expansion for both E. coli and C. difficile on day +1 after reconstitution or infection, respectively, in all groups of mice (Fig. 2B and C). In agreement with our previous findings, the C. difficile burden was significantly reduced by ampicillin treatment in mice reconstituted with mut BLA E. coli but not in mice reconstituted with WT BLA E. coli (Fig. 2D). Direct comparison of the ampicillin-treated mice confirmed a significantly higher burden in mice whose intestinal flora had the capacity to hydrolyze β-lactams (Fig. 2D).
DISCUSSION
The ability of antibiotic-resistant organisms to degrade antibiotics, thus facilitating growth of antibiotic-sensitive bacteria within a microbial community, is well established and has been demonstrated using different antimicrobial compounds (7–9).
In particular, earlier studies in vivo (6, 10–12, 17) revealed that β-lactamase- or chloramphenicol-acetyltransferase-producing bacteria inoculated in conjunction with antibiotic-sensitive pathogens in the hypodermis or lungs could hinder the efficacy of β-lactams or chloramphenicol, respectively. Clinical data also suggested that the presence of one β-lactamase-producing bacterial strain at the site of infection could enhance persistence of a pathogen upon antibiotic treatment (18). In these settings, members of the Bacteroides genus, among the most highly represented genera in the human intestine (19), were also identified as BLA carriers.
Our findings expand on the studies described above by suggesting that ARGs expressed by commensal bacteria can shape the chemical niche of the intestine and confer an apparent antibiotic-resistant phenotype to pathogens in trans, without direct acquisition of ARGs by the pathogenic microbe. We refer to this activity as commensal-mediated pathogen shielding.
Using two different infection models, we show that the production of β-lactamases, a prototypical antibiotic resistance factor, by resident intestinal microbes can significantly reduce the effectiveness of ampicillin treatment, thereby generating a safe environment in which otherwise sensitive pathogens are shielded from this drug.
Although our findings were partly obtained using a model of GF mouse monocolonization, previous studies in healthy volunteers demonstrated that upon treatment with cephalosporins, subjects harboring BLA-producing commensal strains, unlike BLA-negative subjects, had undetectable concentrations of the drug in the feces and maintained a rich microbiota, providing evidence that BLA concentrations sufficient to inactivate antibiotics are commonly achieved in humans (20, 21). Consistent with these observations, our laboratory recently showed that a few bacterial strains, out of the dozens composing the microbiota of a mouse colony treated with ampicillin for over 8 years, had the capacity to hydrolyze ampicillin, while the other bacterial strains, in isolation, remained sensitive to ampicillin and thus were protected in trans by a minor subset of the microbiota (7; see Fig. 4A in reference 22).
Whether or not ARG enrichment within the gut microbiota is detrimental to host health is a complex question, and the answer is likely to be context dependent. For instance, oral administration of recombinant beta-lactamase or BLA-producing bacteria was shown to preserve the integrity of the microbiota following parenteral administration of β-lactam antibiotics in animal models, without affecting drug concentration in the serum (23–28). These approaches were shown to be advantageous in that they preserved colonization resistance against pathogens (23–28).
However, our study suggests that the presence or absence of commensal bacterial strains that inactivate β-lactam antibiotics is likely to impact clinical responses to antibiotic treatment, possibly contributing to interindividual variability in therapy outcomes.
In conclusion, we propose that commensal-mediated pathogen shielding in the intestine can impair the effectiveness of some antibiotic treatments during infection. Although pharmacokinetic studies have generally focused on antibiotic absorption, distribution, enzymatic modification, protein binding, and biliary/renal clearance, the role of microbiota-mediated antibiotic degradation in the gut lumen and its potential for dramatically impacting responses to antibiotic treatment have received less attention. Our findings extend the recently uncovered broad capacity of the gut microbiota to metabolize drugs, affecting their efficacy (29, 30). Within this model, antibiotics represent an additional class of xenobiotics that commensals can metabolize.
We propose that occurrence of pathogen shielding might be a relevant element to consider in the engineering of probiotic bacterial strains to be employed in clinical practice.
MATERIALS AND METHODS
Mouse husbandry.
All experiments using WT mice were performed with C57BL/6J female mice that were 6 to 8 weeks old; the mice were purchased from Jackson Laboratories. Germfree (GF) mice were bred in-house in germfree isolators. After reconstitution, the mice were housed in sterile, autoclaved cages with irradiated food and acidified, autoclaved water. All animals were maintained in a specific-pathogen-free facility at Memorial Sloan Kettering Cancer Center Animal Resource Center. Experiments were performed in compliance with Memorial Sloan-Kettering Cancer Center institutional guidelines and approved by the institution’s Institutional Animal Care and Use Committee.
Generation of E. coli strains.
Plasmids encoding WT TEM-1 β-lactamase (pDIMC8-TEM1) or mutated TEM-1 β-lactamase (pDIMC8-TEM1 W208G) were extracted from the RH06 and RH09 E. coli strains, published elsewhere (13), gel purified, and utilized for the transformation of Stellar competent cells (TaKaRa Bio) according to the manufacturer’s instructions. The resulting strains were utilized for experiments throughout this study. Of note, the plasmids conferred resistance to chloramphenicol, and although the expression of the TEM-1 gene was placed under the regulation of a tac promoter, we did not induce it by IPTG (isopropyl-β-d-thiogalactopyranoside) treatment but rather exclusively relied on leaky transcription of the gene to produce more physiologically relevant conditions.
Antibiotic treatment, reconstitution, and infections.
GF mice were gavaged with either of two strains of E. coli, encoding a functional or a point-mutated version of TEM-1 β-lactamase, respectively. At 1 week after reconstitution, mice were gavaged with 109 CFU of L. monocytogenes strain 10403s and administered 1 mg of ampicillin (Fisher) by oral gavage daily for 2 consecutive days. Animals were euthanized at day 3 postinfection. Upon reconstitution of WT mice with E. coli strains for in vitro experiments involving dilution of cecal content, the mice were treated for 3 days with metronidazole and vancomycin in drinking water (0.5 g/liter), left on regular water for 1 day, and then gavaged with the appropriate E. coli strain. At 1 week after reconstitution, the mice were treated with ampicillin in drinking water (0.5 g/liter) for 2 days prior to being euthanized.
For C. difficile infection experiments, WT C57BL/6 mice were administered a combination of metronidazole, neomycin, and vancomycin (0.25 g/liter each) in drinking water for 3 days, and at 24 h after antibiotic regimen cessation they were injected intraperitoneally with clindamycin (200 μg). On the following day, the mice were reconstituted with either WT or mut BLA E. coli (5 × 104 CFU) and 200 to 500 spores of C. difficile strain VPI 10463 (ATCC 43255).
β-Lactamase detection assay.
Fecal pellets from animals were collected and resuspended in phosphate-buffered saline (PBS) at 100 mg/ml. Samples were left undisturbed for 5 min to allow particulate matter to sediment. Next, 50 μl of the suspension was pipetted into a 96-well plate with 50 μl of nitrocefin (Oxoid), followed by incubation for 30 min at room temperature while protected from light.
CFU enumeration and selective plating.
L. monocytogenes was identified through plating of serial dilutions of homogenized organs (prepared as described elsewhere [14]) or fecal material (resuspended 100 mg/ml in PBS) onto brain heart infusion (BHI) plates supplemented with streptomycin (100 μg/ml) and nalidixic acid (50 μg/ml).
E. coli CFU were enumerated after plating of serial dilution of fecal material onto Luria-Bertani (LB) plates supplemented with chloramphenicol (50 μg/ml). E. coli CFU numbers obtained from plating of ex-GF mice at day of infection onto LB plates (not supplemented with antibiotics) yielded identical numbers, indicating that plasmids carrying the chloramphenicol (CM) resistance cassette, as well as the WT/mut BLA gene, were maintained even in the absence of any selective pressure.
For detection of C. difficile, fecal pellets or cecal content was resuspended in deoxygenated PBS, and 10-fold dilutions were plated on BHI agar supplemented with yeast extract, taurocholate, l-cysteine, cycloserine, and cefoxitin at 37°C in an anaerobic chamber (Coylabs) overnight.
L. monocytogenes culture in cecal content.
Cecal contents were recovered from E. coli reconstituted WT or GF animals, resuspended in PBS at 300 mg/ml (WT), and spun down at 3,000 rpm for 10 s. Serial 1:2 dilutions of the resulting supernatant were generated using PBS, and 100 μl of each dilution was plated in replicate in flat-bottom 96-well plates. An equal volume of BHI medium supplemented with streptomycin (200 μg/ml) and nalidixic acid (100 μg/ml) acid (to prevent growth of residual E. coli) containing 100 to 1,000 CFU of L. monocytogenes 10403s was added on top. L. monocytogenes for this assay was prepared by reinoculating an overnight culture in liquid BHI at 37°C on shaker until the logarithmic phase of growth was reached (optical density [OD] = 0.1 to 0.4). After an overnight incubation at 37°C, the plate was assayed by OD600 reading, and individual dilutions were plated onto BHI-Strep-NA plates to assess L. monocytogenes growth. Normalized inhibition index was calculated as 1/first dilution allowing for L. monocytogenes growth, with the initial dilution being 1:2 to take into account the addition of a volume of BHI equivalent to that of the medium. For example, if the first dilution where L. monocytogenes was detected was 1:16, the resulting inhibition index would be 16. Within each experiment samples were then normalized to the baseline, obtained by averaging the values obtained in the control group, represented by mice reconstituted with mut BLA E. coli.
PCR.
PCR was carried out with specific primers for β-lactamase (forward, 5′-GCTATGTGGCGCGGTATTAT-3′; reverse, 5′-AAGTAAGTTGGCCGCAGTGT-3′; product, 191 bp) and p60 (forward, 5′-GCGCAACAAACTGAAGCAAAGGATGC-3′; reverse, 5′-CTCGCGTTACCAGGCAAATAGATGGACG-3′; product, 1,300 bp) using SapphireAMp Fast PCR master mix (TaKaRa Bio) and the following conditions: 94°C for 1 min, followed by 30 cycles at 98°C for 5 min, 58°C for 5 min, and 72°C for 15 min.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ying Taur and Peter McKenney for critical discussion of the manuscript. RH06 and RH09 E. coli strains carrying the pDIMC8-TEM1 and pDIMC8-TEM1 W208G plasmids were kindly provided by Marc Ostermeier (Johns Hopkins University).
This study was supported by National Institutes of Health grant P30 CA008748 (to Memorial Sloan Kettering Cancer Center), a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award (to T.M.H.) and the NIH grants R01 AI042135 and U01 AI124275 (to E.G.P.). S.B. was supported by an Early Postdoc Mobility Fellowship from the Swiss National Science Foundation (P2EZP3_159083) and an Irvington Fellowship from the Cancer Research Institute (no. 49679).
E.G.P. has received speaker honoraria from Bristol Myers Squibb, Celgene, Seres Therapeutics, MedImmune, Novartis, and Ferring Pharmaceuticals, is an inventor on patent applications WPO2015179437A1 (“Methods and Compositions for Reducing Clostridium difficile Infection”) and WO2017091753A1 (“Methods and Compositions for Reducing Vancomycin-Resistant Enterococci Infection or Colonization”), and holds patents that receive royalties from Seres Therapeutics, Inc.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Nikaido H. 2009. Multidrug resistance in bacteria. Annu Rev Biochem 78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rolain JM. 2013. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front Microbiol 4:173. doi: 10.3389/fmicb.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jernberg C, Lofmark S, Edlund C, Jansson JK. 2007. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 4.Becattini S, Taur Y, Pamer EG. 2016. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. 2012. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorg RA, Lin L, van Doorn GS, Sorg M, Olson J, Nizet V, Veening JW. 2016. Collective resistance in microbial communities by intracellular antibiotic deactivation. PLoS Biol 14:e2000631. doi: 10.1371/journal.pbio.2000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelsic ED, Zhao J, Vetsigian K, Kishony R. 2015. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature 521:516–519. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medaney F, Dimitriu T, Ellis RJ, Raymond B. 2016. Live to cheat another day: bacterial dormancy facilitates the social exploitation of beta-lactamases. ISME J 10:778–787. doi: 10.1038/ismej.2015.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dugatkin LA, Perlin M, Lucas JS, Atlas R. 2005. Group-beneficial traits, frequency-dependent selection and genotypic diversity: an antibiotic resistance paradigm. Proc Biol Sci 272:79–83. doi: 10.1098/rspb.2004.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brook I, Pazzaglia G, Coolbaugh JC, Walker RI. 1983. In-vivo protection of group A beta-haemolytic streptococci from penicillin by beta-lactamase-producing Bacteroides species. J Antimicrob Chemother 12:599–606. doi: 10.1093/jac/12.6.599. [DOI] [PubMed] [Google Scholar]
- 11.Brook I, Pazzaglia G, Coolbaugh JC, Walker RI. 1984. In vivo protection of penicillin-susceptible Bacteroides melaninogenicus from penicillin by facultative bacteria which produce beta-lactamase. Can J Microbiol 30:98–104. doi: 10.1139/m84-017. [DOI] [PubMed] [Google Scholar]
- 12.Hackman AS, Wilkins TD. 1975. In vivo protection of Fusobacterium necrophorum from penicillin by Bacteroides fragilis. Antimicrob Agents Chemother 7:698–703. doi: 10.1128/aac.7.5.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohka T, Heins RA, Phelan RM, Greisler JM, Townsend CA, Ostermeier M. 2009. An externally tunable bacterial band-pass filter. Proc Natl Acad Sci U S A 106:10135–10140. doi: 10.1073/pnas.0901246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becattini S, Littmann ER, Carter RA, Kim SG, Morjaria SM, Ling L, Gyaltshen Y, Fontana E, Taur Y, Leiner IM, Pamer EG. 2017. Commensal microbes provide first line defense against Listeria monocytogenes infection. J Exp Med 214:1973–1989. doi: 10.1084/jem.20170495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pamer EG. 2016. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352:535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brook I. 2009. The role of beta-lactamase-producing-bacteria in mixed infections. BMC Infect Dis 9:202. doi: 10.1186/1471-2334-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brook I. 2004. Beta-lactamase-producing bacteria in mixed infections. Clin Microbiol Infect 10:777–784. doi: 10.1111/j.1198-743X.2004.00962.x. [DOI] [PubMed] [Google Scholar]
- 19.Human Microbiome Project Consortium. 2012. Structure, function, and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonard F, Andremont A, Leclerq B, Labia R, Tancrede C. 1989. Use of beta-lactamase-producing anaerobes to prevent ceftriaxone from degrading intestinal resistance to colonization. J Infect Dis 160:274–280. doi: 10.1093/infdis/160.2.274. [DOI] [PubMed] [Google Scholar]
- 21.Chachaty E, Bourneix C, Renard S, Bonnay M, Andremont A. 1993. Shedding of Clostridium difficile, fecal beta-lactamase activity, and gastrointestinal symptoms in 51 volunteers treated with oral cefixime. Antimicrob Agents Chemother 37:1432–1435. doi: 10.1128/aac.37.7.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caballero S, Kim S, Carter RA, Leiner IM, Susac B, Miller L, Kim GJ, Ling L, Pamer EG. 2017. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe 21:592–602. doi: 10.1016/j.chom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebhart D, Lok S, Clare S, Tomas M, Stares M, Scholl D, Donskey CJ, Lawley TD, Govoni GR. 2015. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio 6:e02368-14. doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stiefel U, Nerandzic MM, Pultz MJ, Donskey CJ. 2014. Gastrointestinal colonization with a cephalosporinase-producing bacteroides species preserves colonization resistance against vancomycin-resistant enterococcus and Clostridium difficile in cephalosporin-treated mice. Antimicrob Agents Chemother 58:4535–4542. doi: 10.1128/AAC.02782-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarkkanen AM, Heinonen T, Jogi R, Mentula S, van der Rest ME, Donskey CJ, Kemppainen T, Gurbanov K, Nord CE. 2009. P1A recombinant beta-lactamase prevents emergence of antimicrobial resistance in gut microflora of healthy subjects during intravenous administration of ampicillin. Antimicrob Agents Chemother 53:2455–2462. doi: 10.1128/AAC.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiefel U, Harmoinen J, Koski P, Kaariainen S, Wickstrand N, Lindevall K, Pultz NJ, Bonomo RA, Helfand MS, Donskey CJ. 2005. Orally administered recombinant metallo-beta-lactamase preserves colonization resistance of piperacillin-tazobactam-treated mice. Antimicrob Agents Chemother 49:5190–5191. doi: 10.1128/AAC.49.12.5190-5191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harmoinen J, Mentula S, Heikkila M, van der Rest M, Rajala-Schultz PJ, Donskey CJ, Frias R, Koski P, Wickstrand N, Jousimies-Somer H, Westermarck E, Lindevall K. 2004. Orally administered targeted recombinant beta-lactamase prevents ampicillin-induced selective pressure on the gut microbiota: a novel approach to reducing antimicrobial resistance. Antimicrob Agents Chemother 48:75–79. doi: 10.1128/aac.48.1.75-79.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiefel U, Pultz NJ, Harmoinen J, Koski P, Lindevall K, Helfand MS, Donskey CJ. 2003. Oral administration of beta-lactamase preserves colonization resistance of piperacillin-treated mice. J Infect Dis 188:1605–1609. doi: 10.1086/379153. [DOI] [PubMed] [Google Scholar]
- 29.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. 2019. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570:462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. 2019. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 363:eaat9931. doi: 10.1126/science.aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.