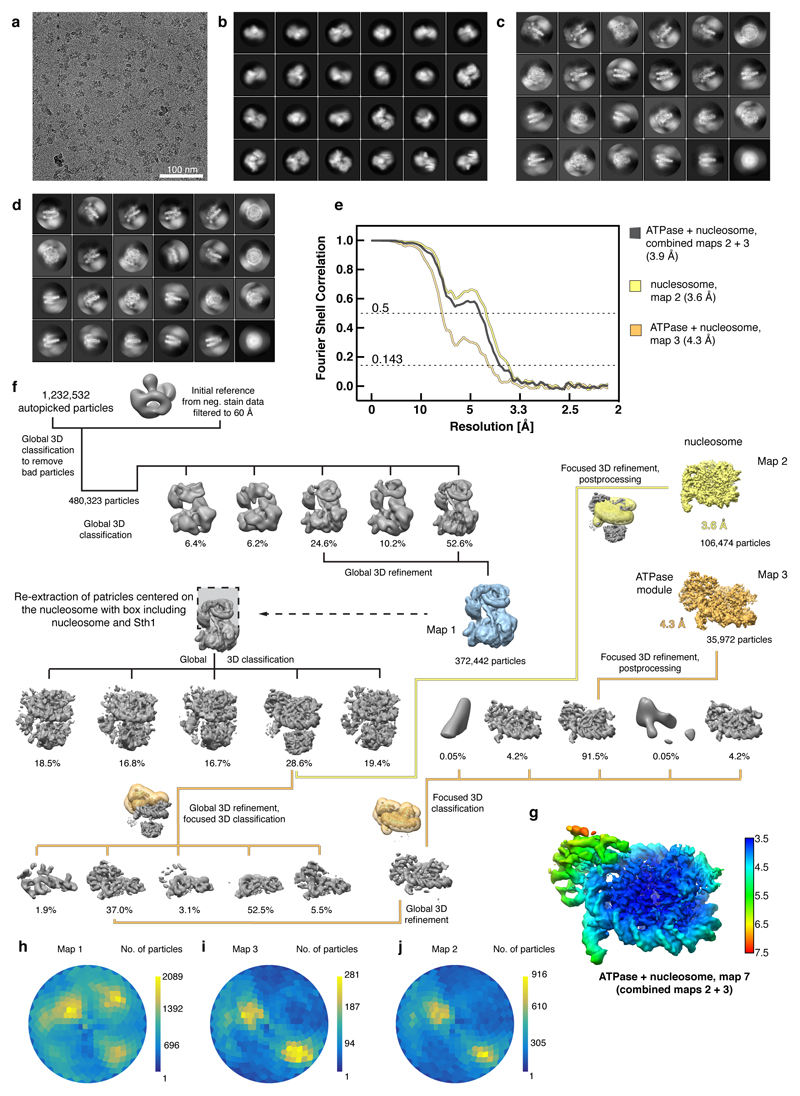
Extended Data Figure 2. Cryo-EM analysis of the RSC-nucleosome complex. Related to Figures 1 – 3.
a. Representative cryo-EM micrograph of the RSC-nucleosome complex shows homogeneously distributed individual particles.
b-d. 2D class averages of the RSC-nucleosome complex (b), the ATPase-nucleosome subcomplex (c) and the nucleosome subcomplex (d).
e. Fourier shell correlation plots reveal the overall resolutions of the cryo-EM reconstructions.
f. Cryo-EM processing workflow for the reconstructions of the RSC-nucleosome complex, the ATPase-nucleosome subcomplex, and the nucleosome subcomplex. Particle distribution after 3D classifications is indicated below the corresponding map. The final maps are shown in colours. The masks used for focused classifications and refinements are colour coded corresponding to the final maps they were used for. Views are generally rotated by 180° with respect to Figure 1c, left.
g. Local resolution estimation of the combined ATPase-nucleosome map as implemented in RELION42. We note that the resolution of the peripheral area with the ATPase module is overestimated.
h-j. Angular distribution plot for all particles contributing to the final reconstructions of the RSC-nucleosome complex (h), the ATPase-nucleosome subcomplex (i) and the nucleosome subcomplex (j).