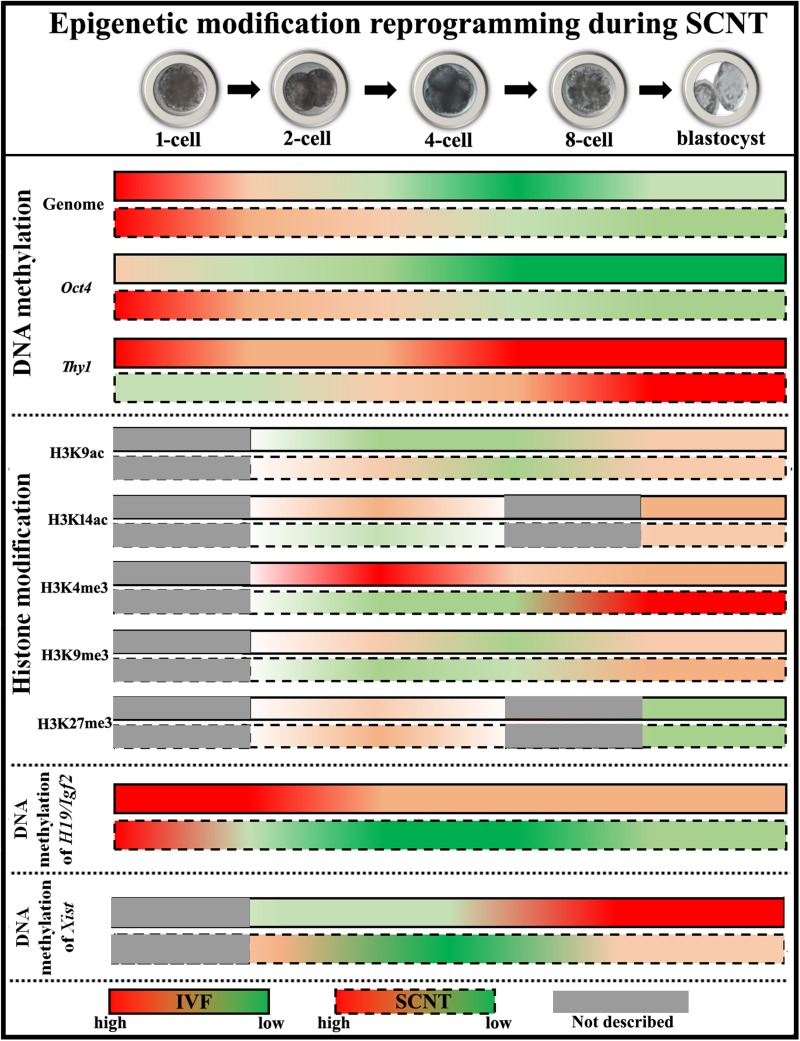
FIGURE 2.
Diagram of epigenetic modification changes during SCNT-mediated nuclear reprogramming. The data of pig embryos are adopted to describe epigenetic modification reprogramming. For DNA methylation, cloned embryos demonstrate delayed DNA demethylation and incomplete DNA remethylation of genome (the data shown here represent DNA methylation status at centromeric repeats, which partly reflects genome DNA status), high DNA methylation status of pluripotency-related gene Oct4, and low DNA methylation levels of tissue-specific gene Thy1, respectively. For histone modifications, low levels of histone acetylation (H3K9ac at the ZGA stage and H3K14ac at the blastocyst stage) and H3K4me3, and high levels of histone methylation (H3K9me3 after ZGA and H3K27me3 at the 2-cell stage) are observed in cloned embryos. For genomic imprinting, DNA methylation of H19/Igf2 is not maintained during SCNT. For XCI, DNA methylation of Xist is not fully established in female cloned embryos.