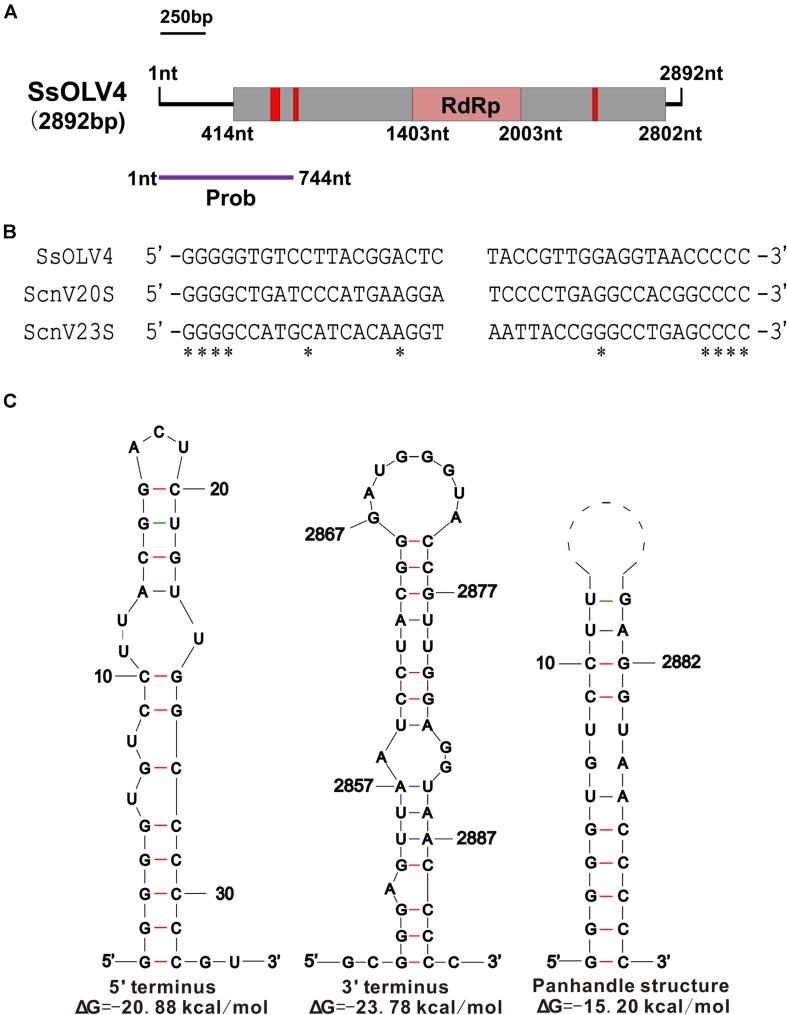
FIGURE 2.
The genome organization and the terminal structure of SsOLV4. (A) A schematic diagram of the genome organization of SsOLV4. SsOLV4 contains a single open reading frame (ORF) shown as a gray box (414–2,802 nt), which putatively encodes the RNA-dependent-RNA polymerase (RdRp) domain, shown as a pink box (1,403–2,003 nt). The predicted nuclear location signal peptide sequences are shown as red boxes. The DNA segment amplified by RT-PCR was used as a probe in northern hybridization experiments and is shown as a purple line. Nt, nucleotide position on the genome. (B) Alignment of the 5′- and 3′-end regions of SsOLV4 and the two yeast narnaviruses ScnV20S (Saccharomyces 20S RNA narnavirus) and ScnV23S (Saccharomyces 23S RNA narnavirus); asterisks indicate the same nucleic acid. (C) Potential secondary structures of the 5′- (left) and 3′-termini (middle) of SsOLV4 and a putative panhandle structure (right) formed by the inverted complementarity at two terminal sequences are depicted; the ΔΔG values (kcal/mol) were calculated using Mfold RNA structure software.