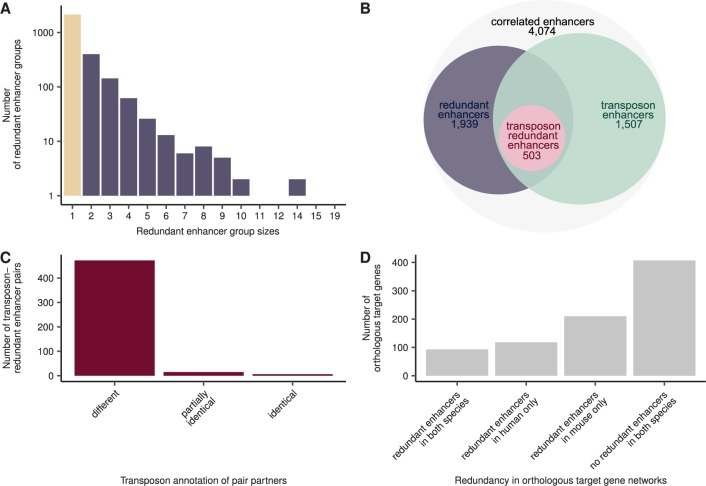
Fig. 3.
—Redundant enhancers in the mouse genome show similar properties to those in the human genome, but redundancy in orthologous regulatory networks is mainly lineage-specific. (A) Redundant enhancers in the mouse genome form groups of different sizes, similar as in human. Depicted are the number of nonredundant enhancers (beige) and redundant-enhancer groups (purple). (B) The scheme depicts the connection of the redundant enhancer, transposon enhancer, and transposon-redundant enhancer sets. See figure 2B for details. (C) The enhancers of redundant enhancer pairs show the same transposon annotation pattern as in human. Depicted are the number of transposon-redundant enhancer pairs where partners have different, partially identical (some transposons are identical), or identical transposon species annotation. (D) A large fraction of target genes that are orthologous between human and mouse are regulated by redundant enhancers in just one of the two species. Shown are the common orthologous enhancer target genes between the human- and mouse-correlated enhancer data sets and their regulation by redundant enhancers in human and mouse.