Table 1.
In vitro hepatic metabolism data for a panel of triazole-based NAMPT inhibitors.
| Compound | Structure | [M+H]+ | Metabolic stabilitya | Revealed metabolic pathwaysb | |||||
|---|---|---|---|---|---|---|---|---|---|
| RLM | HLM | Pyridine N-oxidation |
Aromatic oxidation |
Aliphatic oxidation | Hydrolysis | Other | |||
| A1 |  |
441 | 51 | 72 | ● | ◌ | ● | ◌ | |
| A2 | 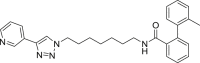 |
454 | 30 | 84 | ● | ◌ | ● | ◌ | |
| A3 | 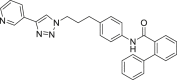 |
460 | 21 | 78 | ● | ● | ● | ◌ | |
| A4 | 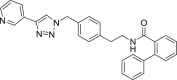 |
460 | 46 | 91 | ● | ● | ● | ◌ | |
| A5 |  |
426 | 62 | 92 | ● | ● | ● | ◌ | |
| A6 |  |
440 | 45 | 88 | ● | ● | ● | ◌ | Oxidative N-dealkylation |
| E1 | 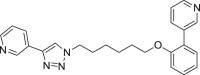 |
400 | 23 | 60 | ● | ● | ◌ | ◌ | |
| E2 |  |
413 | 8 | 32 | ◌ | ● | ● | ◌ | |
| E3 |  |
433 | 70 | 94 | ● | ● | ● | ◌ | |
| E4 | 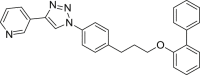 |
433 | 77 | >99 | ◌ | ● | ● | ◌ | |
| E5 | 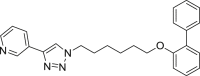 |
399 | 31 | 49 | ● | ● | ● | ◌ | |
| E6 | 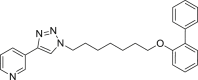 |
413 | 14 | 61 | ● | ● | ● | ◌ | |
| E7 |  |
427 | 16 | 84 | ● | ● | ● | ◌ | |
| C1 |  |
442 | 66 (61)c | 67 (87)c | ● | ● | ● | ● | |
| C2 |  |
476 | 69 (92)c | 90 | ● | ● | ◌ | ● | |
| C3 |  |
456 | 60 (89)c | 86 (92)c | ● | ● | ● | ● | |
| U1 |  |
441 | 79 | 95 | ● | ● | ● | ◌ | |
| U2 |  |
455 | 60 | 85 | ● | ● | ● | ◌ | |
| U3 | 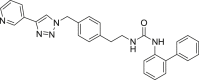 |
475 | 95 | 96 | ● | ● | ● | ◌ | |
| S1 | 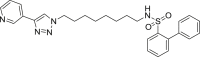 |
490 | 62 | 78 | ● | ● | ● | ◌ | Oxidative N-dealkylation |
| S2 | 499 | 45 | 80 | ● | ◌ | ● | ◌ | Oxidative N-dealkylation |
|
| S3 | 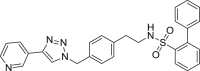 |
496 | 76 | 84 | ● | ◌ | ● | ◌ | Oxidative N-dealkylation |
| N1 |  |
446 | 38 | 74 | ● | ◌ | ● | ◌ | Oxidative N-dealkylation |
| T1 |  |
464 | 47 | 87 | ● | ● | ● | ◌ | |
The metabolic stability was assessed by incubating each compound at 50 μM concentration with rat (RLM) or human (HLM) liver microsomes in the presence or absence of NADPH-regenerating system [4]. Residual substrate percentage was evaluated by LC-UV analysis measuring the remaining substrate after incubation.
For structural characterization of the metabolites, all samples were analyzed by LC-ESI-MSn.
Residual substrate in incubations without the presence of NADPH-regenerating system.
