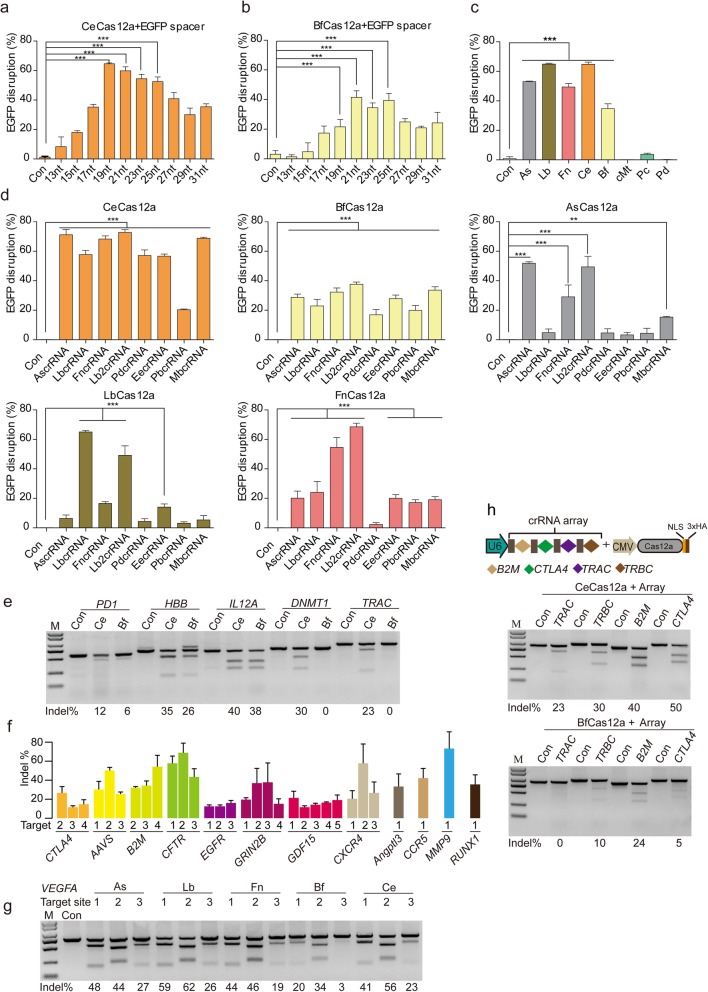
Fig. 2.
CeCas12a and BfCas12a mediate gene editing in human cells. a Efficiencies of EGFP disruption in human cells mediated by CeCas12a and crRNAs bearing variable-length complementarity regions for the target site of EGFP in human cells. Error bars indicate standard errors of means (s.e.m.), n = 3. ***P < 0.001 (Mann-Whitney). b Efficiencies of EGFP disruption mediated by BfCas12a and crRNAs bearing variable-length complementarity regions for the target site of EGFP in human cells. Error bars represent s.e.m., n = 3. ***P < 0.001 (Mann-Whitney). c Efficiencies of EGFP disruption in human cells mediated by Cas12a orthologs and crRNA bearing 23-nt length complementarity regions for the target site of EGFP in human cells. Error bars represent s.e.m., n = 3. ***P < 0.001 (Mann-Whitney). d The suitability of Cas12a with crRNAs containing different loop regions from Cas12a orthologs. Data are shown as mean ± s.e.m. (n = 3); **P < 0.01, ***P < 0.001 (Mann-Whitney). e Efficiencies of targeted indel mutations introduced at five different human endogenous gene targets by CeCas12a and BfCas12a. Indel frequencies of PD1, HBB, IL12A, DNMT1, and TRAC were measured by T7E1 assay. f Summaries of the activities of CeCas12a at TTTV PAMs at a diverse panel of target sites in HEK293T cells. For indel percentages, each column represents the mean of n = 3 transfected cell cultures. Indel frequencies were analyzed by T7E1 assay. g Comparison of CeCas12a and Cas12a orthologs’ gene-editing efficiencies. Three target sites are from VEGFA loci. Indel frequencies analyzed by T7E1 assay. h Cas12a-mediated multiplex gene editing in human cells. Schematic of CRISPR array construct with the U6 direct repeat spacer cassette containing B2M, CTLA4, TRAC, and TRBC protospacer sequences (top). Indel frequencies were analyzed by T7E1 assay after transfection with CRISPR array and Cas12a plasmids (bottom)