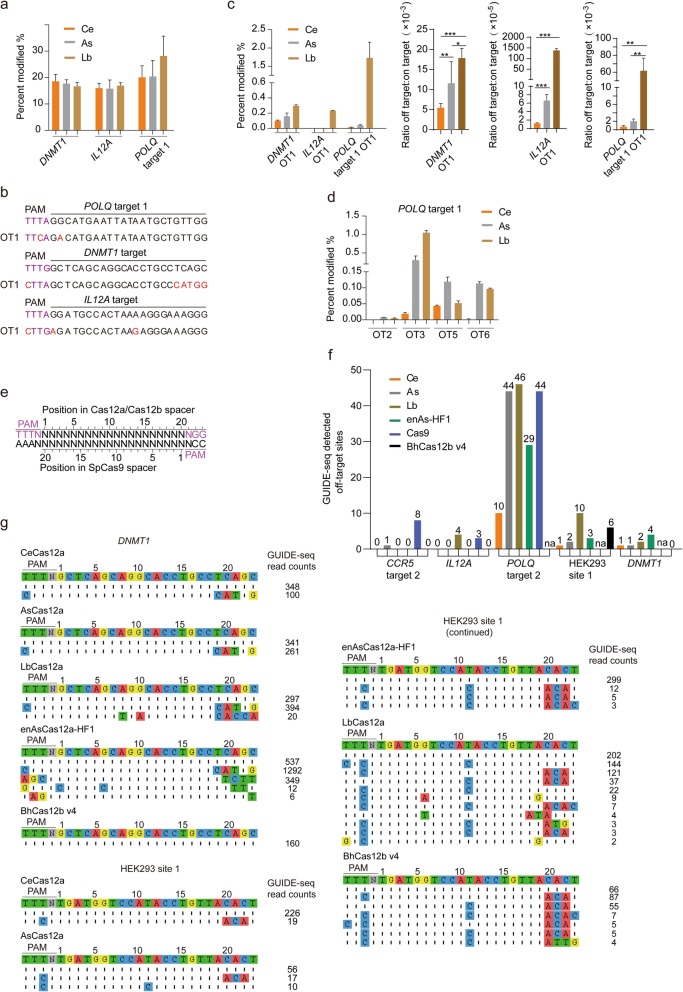
Fig. 4.
Assessment off-target effects of CRISPR-Cas nucleases. a Mean percent modification by Lb, As, and Ce at POLQ, DNMT1, and IL12A on-target sites. Percent modifications were determined by deep sequencing. Bars show mean ± s.e.m. for n = 3 transfected cell cultures. Indel mutation frequencies were indicated by the percentage of reads containing indels compared with the wild-type sequence reads. b Illustration of off-target sites at PAM sequence. Mismatches at PAM sequence and spacer are red. c Mean percent modification at off-target site 1 (OT1) (left), and specificity ratios of LbCas12a, AsCas12a, and CeCas12a with the POLQ, DNMT1, and IL12A crRNAs, plotted as the ratio of OT1 site activity to on-target activity (right). Data are shown as mean ± s.e.m. (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 (Mann-Whitney). d Mean percent modification by Lb, As, and Ce at off-target sites when targeting POLQ target 1. OT2, OT3, OT5, and OT6 were predicted by Cas-OFFinder, and mismatches are in spacer region (Additional file 1: Figure S13). e Matched target sites for Cas12a, Cas12b, and SpCas9 that share a common protospacer sequence. f Histograms illustrating the number of GUIDE-seq detected off-target sites for CeCas12a, AsCas12a, LbCas12a, enAsCas12a-HF1, spCas9, and BhCas12b v4. na, not assessed. g Off-target sites for CeCas12a, AsCas12a, LbCas12a, enAsCas12a-HF1, and BhCas12b v4 with DNMT1 and HEK293 site1 crRNAs, determined using GUIDE-seq in HEK293 cells. Mismatched positions are highlighted in color, and GUIDE-seq read counts are shown to the right of the on- or off-target sequences