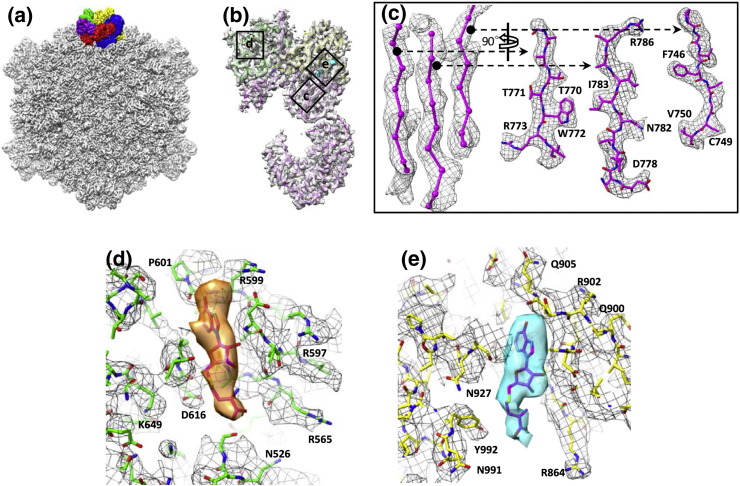
Fig. 1.
Structure of transcribing cypovirus. (a) Overall structure of a transcribing cypovirus filtered at 6 Å resolution along a 2-fold axis. The five VP3 monomers at a fivefold vertex are shown in different colors. (b) Transparent views of the density map of VP3 with its atomic model (only backbone shown) superimposed. The models of the MTase-1 and MTase-2 domains are in green and yellow, respectively. Positions of views labeled c, d, and e are shown in (c), (d), and (e). (c) Zoom-in view of a β-sheet segmented from the structure of VP3 with its atomic model superimposed. (d) Zoom-in view of the SAM/SAH binding site in the MTase-1 domain. An atomic model of SAH is fitted into the density (orange) that is attributed to SAM/SAH molecules located at the MTase-1 domain. Atomic model (sticks) superimposed on its density map (mesh) with some side chains labeled. (e) Zoom-in view of the SAM/SAH binding site of the MTase-2 domain. An atomic model of SAH is fitted in the structure (cyan) that is attributed to SAM/SAH molecules located at the MTase-2 domain. Atomic model (sticks) superimposed on its density map (mesh) with a number of side chains labeled.