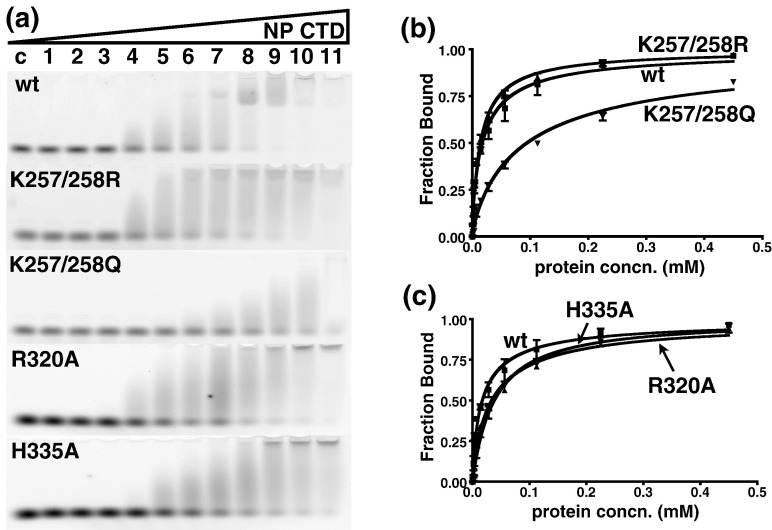
Fig. 8.
EMSA of SARS-CoV NP CTD mutants. (a) Mobility shift of dT20 bound to wild-type (wt), K257R/K258R (K257/258R), K257Q/K258Q (K257/258Q), R320A, and H335A mutant proteins. The protein concentration was increased by a factor of 2, starting from lane 1 (439 nM) to lane 11 (0.45 mM). Lane C, negative control. (b) Binding curve of the K257/K258 double mutant towards dT20, compared to that of the wild-type protein. (c) Binding curve of the R320A and H335A mutants towards dT20, compared to that of the wild-type protein. Each curve in (b) and (c) represents the best fit from three independent assays. Results are summarized in Table 2.